CX-revision
Renee Matthews
2025-11-03
Last updated: 2025-11-03
Checks: 7 0
Knit directory: CX_revison/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20251031) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d642f3e. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Unstaged changes:
Modified: CX_revison.Rproj
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/CX-revision.Rmd) and HTML
(docs/CX-revision.html) files. If you’ve configured a
remote Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d642f3e | reneeisnowhere | 2025-11-03 | wflow_publish("analysis/CX-revision.Rmd") |
| html | f31ecb6 | reneeisnowhere | 2025-10-31 | Build site. |
| Rmd | 45f92fd | reneeisnowhere | 2025-10-31 | update links |
| html | 10b921a | reneeisnowhere | 2025-10-31 | Build site. |
| Rmd | 9928965 | reneeisnowhere | 2025-10-31 | updated |
| html | f2ba629 | reneeisnowhere | 2025-10-31 | Build site. |
| Rmd | 635aef2 | reneeisnowhere | 2025-10-31 | wflow_publish("analysis/CX-revision.Rmd") |
library(tidyverse)
library(readxl)
library(readr)
library(drc)adding input data
X87_1_0hr_ipsc_96wellplate_baseline_no_treatment_10_14_2025_251014 <- read_excel("data/Presto_blue_results/87-1_0hr_ipsc_96wellplate_baseline_no_treatment_10-14-2025_251014.xls",
skip = 34)
X87_1_48hr_ipsc_96wellplate_CX_DOX_Veh_10_16_2025_251016 <- read_excel("data/Presto_blue_results/87-1_48hr_ipsc_96wellplate_CX-DOX-Veh_10-16-2025_251016.xls",
skip = 34)
combo_time_data <-
X87_1_0hr_ipsc_96wellplate_baseline_no_treatment_10_14_2025_251014 %>%
dplyr::filter(Well=="Read 2:560,590") %>%
mutate(Well="zero") %>%
bind_rows(X87_1_48hr_ipsc_96wellplate_CX_DOX_Veh_10_16_2025_251016 %>%
dplyr::filter(Well=="Read 2:560,590") %>% mutate(Well="two_day")) %>%
dplyr::rename("Time"=Well) %>%
pivot_longer(., A1:H12, names_to = "Well", values_to = "reading") %>%
pivot_wider(id_cols=c("Well"), names_from = "Time", values_from = "reading")Plate_map <- read_excel("data/Presto_blue_results/Plate_map.xlsx") base_df <- Plate_map %>%
pivot_longer(cols = everything(), names_to = "Well") %>%
separate(col = value, into = c("trt", "conc"), sep = "_") %>%
left_join(., combo_time_data, by = "Well") %>%
pivot_longer(cols = c(zero, two_day), names_to = "Time") %>%
mutate(
trt = factor(trt, levels = c("CX", "DOX", "VEH", "Unt", "blank")), # check these levels match your data
conc = factor(conc, levels = c("0","0.1","0.5","1","5","10","50","blank")),
Time = factor(Time, levels = c("zero","two_day"))
)
# 1. subtract blank per Time and clip negatives
processed <- base_df %>%
group_by(Time) %>%
mutate(
blank_mean = mean(value[trt == "blank"], na.rm = TRUE),
adj_reading = pmax(value - blank_mean, 0)
) %>%
ungroup()
untx_per_ct <- processed %>%
filter(trt == "Unt") %>% # only VEH rows
group_by(conc, Time) %>%
summarise(
unt_mean = mean(adj_reading, na.rm = TRUE),
n_unt = n(),
.groups = "drop"
)
with_veh <-processed %>%
left_join(untx_per_ct, by = c( "Time")) %>%
mutate(
norm_to_veh = case_when(
is.na(unt_mean) ~ NA_real_, # no VEH present for that conc×Time
unt_mean == 0 ~ NA_real_, # avoid division by zero
TRUE ~ (adj_reading / unt_mean) * 100
)
)
wide_norm <-
with_veh %>%
dplyr::select(Well, trt, conc.x, Time, norm_to_veh) %>%
pivot_wider(id_cols = c(Well, trt, conc.x), names_from = Time, values_from = norm_to_veh)
# Step 5: compute between-time ratio (two_day relative to zero) safely
final_df <- wide_norm %>%
mutate(
hr_norm = case_when(
is.na(zero) | is.na(two_day) ~ NA_real_,
zero == 0 ~ NA_real_, # choose NA to flag unreliable ratio (or 0 if you prefer)
TRUE ~ (two_day / zero) * 100
)
) %>%
filter(trt != "blank")final_df %>%
dplyr::rename("conc"=conc.x) %>%
ggplot(., aes(x=conc, y=zero))+
geom_boxplot(aes(fill=trt))+
theme_bw()+
ggtitle("Box plot of normalized day zero across treatments")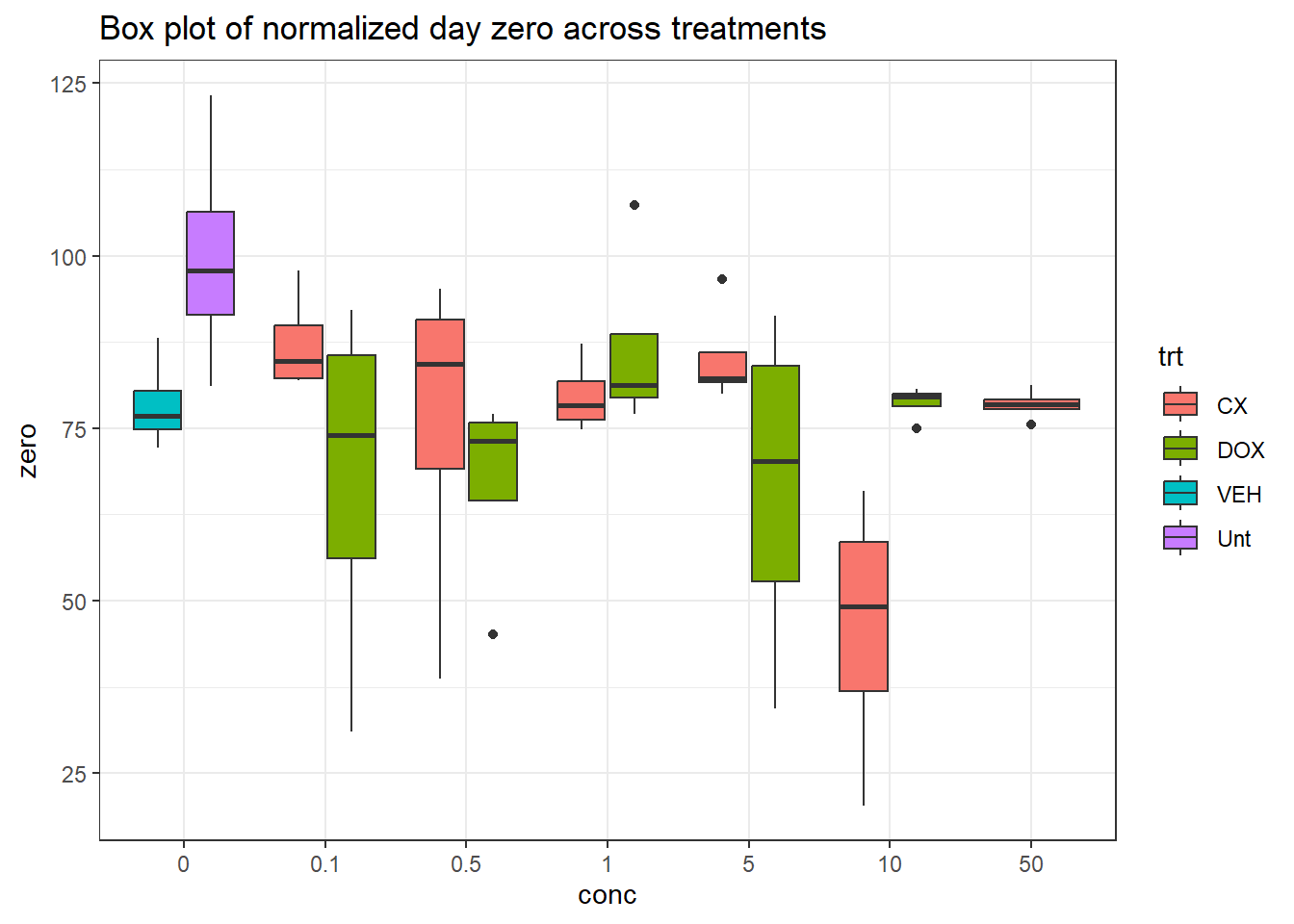
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
final_df %>%
dplyr::rename("conc"=conc.x) %>%
ggplot(., aes(x=conc, y=two_day))+
geom_boxplot(aes(fill=trt))+
theme_bw()+
ggtitle("Box plot of normalized day two across treatments")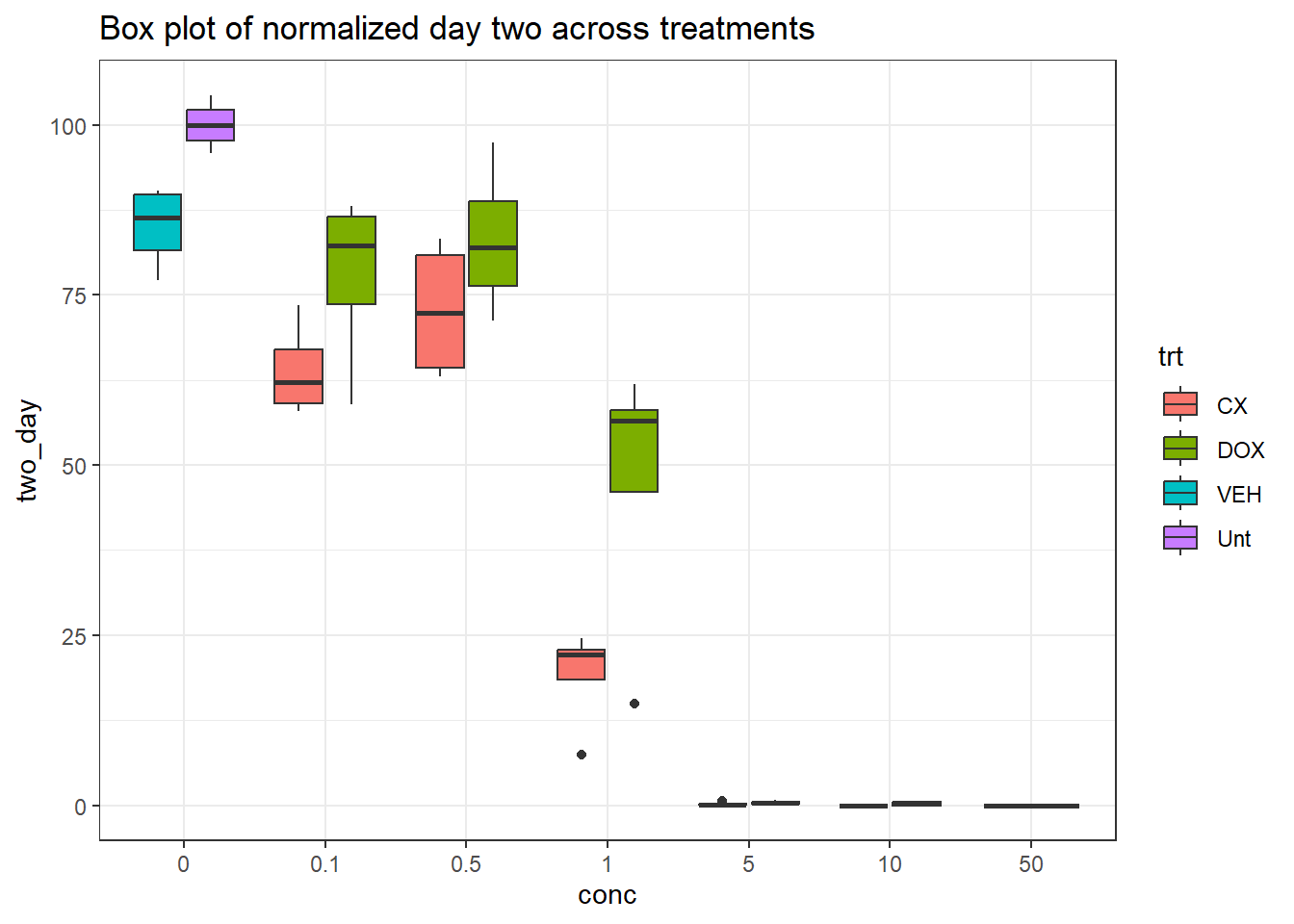
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
final_df %>%
dplyr::rename("conc"=conc.x) %>%
ggplot(., aes(x=conc, y=hr_norm))+
geom_boxplot(aes(fill=trt))+
theme_bw()+
ggtitle("Box plot of normalized across time for each well (between plates)")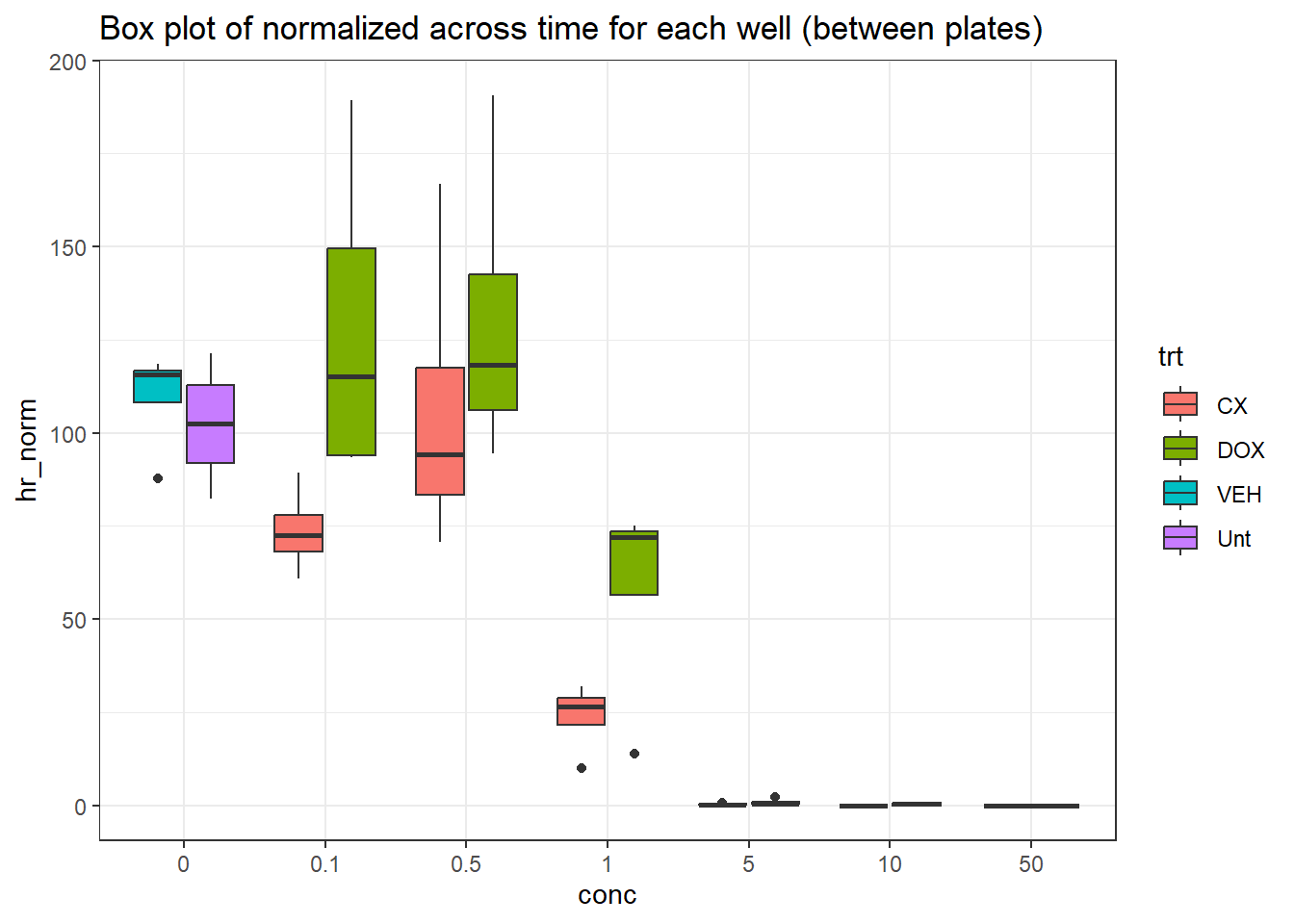
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
# Fit
gpt_try <- final_df %>%
filter(trt == "CX") %>%
mutate(conc.x = as.numeric(as.character(conc.x)))
model <- final_df %>%
filter(trt == "CX") %>%
mutate(conc.x = as.numeric(as.character(conc.x))) %>%
drm(hr_norm ~ conc.x, data = ., fct = L.4())
# Predict across log-spaced range
pred <- data.frame(conc.x = exp(seq(log(min(gpt_try$conc.x[gpt_try$conc.x > 0])),
log(max(gpt_try$conc.x)),
length.out = 100)))
pred$hr_norm <- predict(model, newdata = pred)
LD50 <- ED(model, 50, interval = "delta") # 50% effective dose
Estimated effective doses
Estimate Std. Error Lower Upper
e:1:50 0.939655 0.077668 0.777642 1.101668LD50_val <- as.numeric(LD50[1,1])
# Plot
gpt_try %>%
filter(trt == "CX") %>%
ggplot(aes(x = conc.x, y = hr_norm)) +
geom_point() +
geom_line(data = pred, aes(x = conc.x, y = hr_norm), color = "blue", size = 1) +
geom_vline(xintercept = LD50_val, color = "red", linetype = "dashed") +
annotate("text", x = LD50_val, y = max(gpt_try$hr_norm, na.rm = TRUE),
label = paste0("LD50 = ", signif(LD50_val, 3)),
color = "red", angle = 90, vjust = -0.5, hjust = 1.1) +
scale_x_log10() +
theme_minimal()+
ggtitle("iPSC 87-1 Dose response Assay for CX")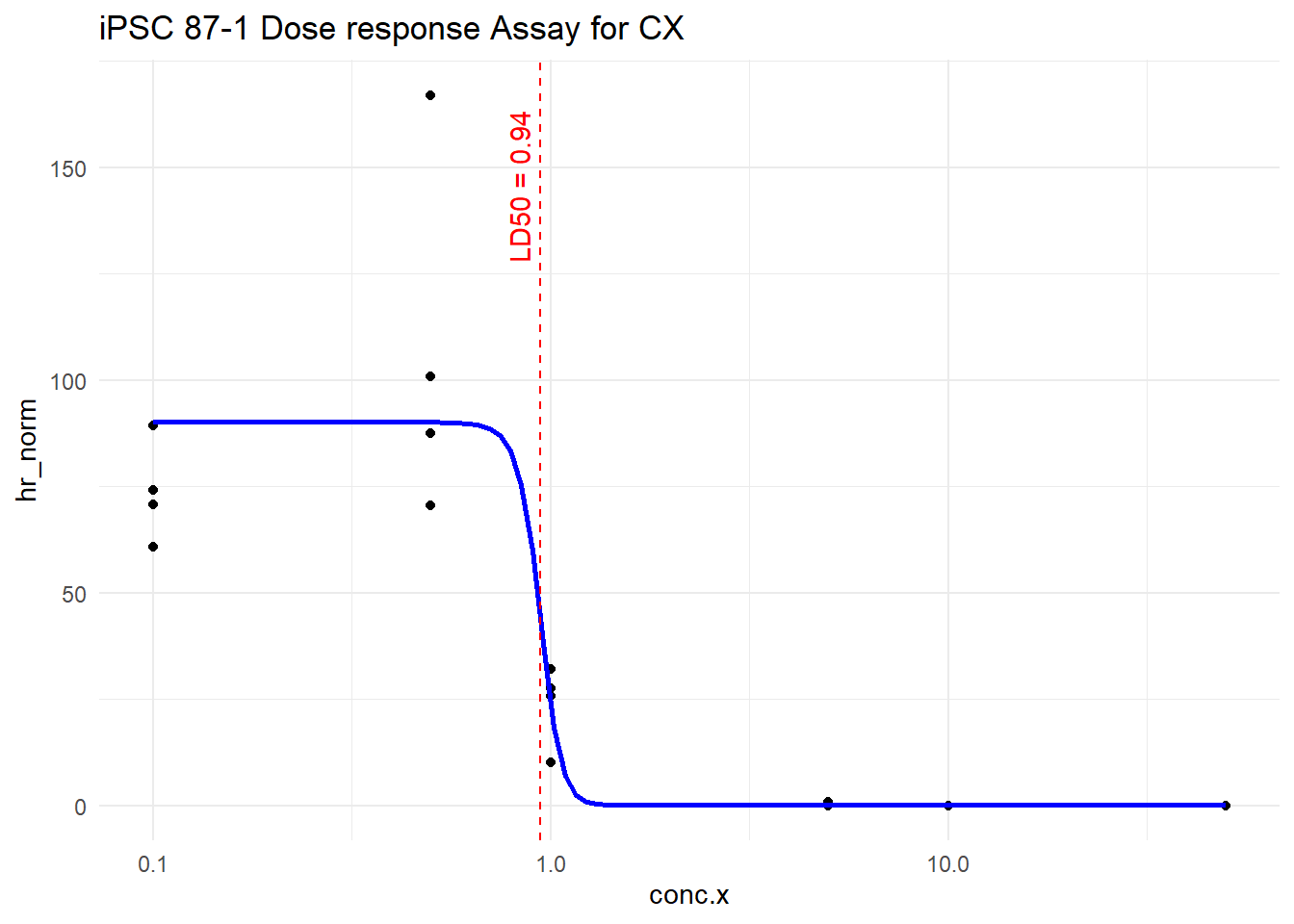
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
# Fit
gpt_try_DOX <- final_df %>%
filter(trt == "DOX") %>%
mutate(conc.x = as.numeric(as.character(conc.x)))
model_DOX <- final_df %>%
filter(trt == "DOX") %>%
mutate(conc.x = as.numeric(as.character(conc.x))) %>%
drm(hr_norm ~ conc.x, data = ., fct = L.4())
# Predict across log-spaced range
pred_DOX <- data.frame(conc.x = exp(seq(log(min(gpt_try_DOX$conc.x[gpt_try_DOX$conc.x > 0])),
log(max(gpt_try_DOX$conc.x)),
length.out = 100)))
pred_DOX$hr_norm <- predict(model_DOX, newdata = pred_DOX)
LD50_DOX <- ED(model_DOX, 50, interval = "delta") # 50% effective dose
Estimated effective doses
Estimate Std. Error Lower Upper
e:1:50 0.97482 0.07383 0.81830 1.13133LD50_val_DOX <- as.numeric(LD50_DOX[1,1])
# Plot
gpt_try_DOX %>%
filter(trt == "DOX") %>%
ggplot(aes(x = conc.x, y = hr_norm)) +
geom_point() +
geom_line(data = pred_DOX, aes(x = conc.x, y = hr_norm), color = "blue", size = 1) +
geom_vline(xintercept = LD50_val_DOX, color = "red", linetype = "dashed") +
annotate("text", x = LD50_val_DOX, y = max(gpt_try_DOX$hr_norm, na.rm = TRUE),
label = paste0("LD50 = ", signif(LD50_val_DOX, 3)),
color = "red", angle = 90, vjust = -0.5, hjust = 1.1) +
scale_x_log10() +
theme_minimal()+
ggtitle("iPSC 87-1 Dose response Assay for DOX")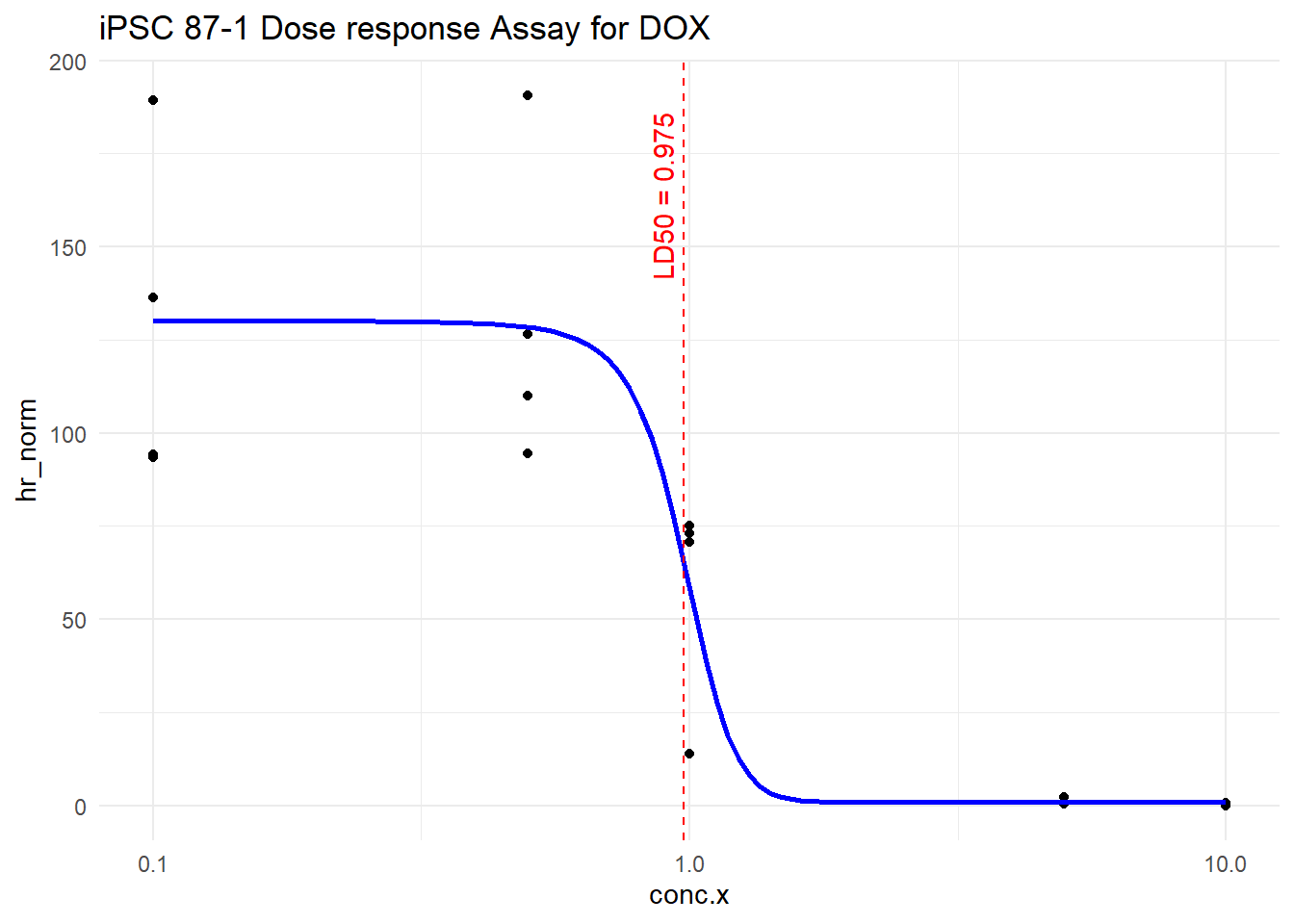
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
75-1 iPSC renee
T0_75_iPSC_p1_viability <- read_excel("data/Presto_blue_results/75_p1_iPSC_T0_251026.xls",
skip = 26)
T0_75_iPSC_p2_viability <- read_excel("data/Presto_blue_results/75_p2_iPSC_T0_251026.xls",
skip = 26)
T48_75_iPSC_p1_viability <- read_excel("data/Presto_blue_results/75_iPSC_p1_T48_251028.xlsx",
skip = 26)
T48_75_iPSC_p2_viability <- read_excel("data/Presto_blue_results/75_iPSC_p2_T48_251028.xlsx",
skip = 26)
iPSC_75_viability_data <-
T0_75_iPSC_p1_viability %>%
mutate(Well="T0_p1") %>%
bind_rows(T0_75_iPSC_p2_viability %>%
mutate(Well="T0_p2")) %>%
bind_rows(T48_75_iPSC_p1_viability %>%
mutate(Well="T48_p1")) %>%
bind_rows(T48_75_iPSC_p2_viability %>%
mutate(Well="T48_p2")) %>%
dplyr::rename("Time_Plate"=Well) %>%
pivot_longer(., A1:H12, names_to = "Well", values_to = "reading") %>%
pivot_wider(id_cols=c("Well"), names_from = "Time_Plate", values_from = "reading")iPSC_plate_map <- read_excel("data/Presto_blue_results/iPSC_cell_viability_platemap.xlsx",
skip = 9) base_df_75 <-iPSC_plate_map %>%
separate(tx_conc, into = c("tx","conc"), sep = "_") %>%
left_join(., iPSC_75_viability_data, by = "Well") %>%
pivot_longer(cols = c(T0_p1, T0_p2, T48_p1,T48_p2), names_to = "Time_plate") %>%
separate(Time_plate, into = c("Time","plate"), sep = "_") %>%
mutate(
tx = factor(tx, levels = c("CX", "DOX", "VEH", "untx", "blank")), # check these levels match your data
conc = factor(conc, levels = c("0","0.05","0.1","0.5","0.75","1","5","10","na")),
Time = factor(Time, levels = c("T0","T48"))
)
# 1. subtract blank per Time and clip negatives
processed_75 <- base_df_75 %>%
group_by(plate,Time) %>%
mutate(
blank_mean = mean(value[tx == "blank"], na.rm = TRUE),
adj_reading = pmax(value - blank_mean, 0)
) %>%
ungroup()
untx_per_plate_75 <- processed_75 %>%
filter(tx == "untx") %>%
group_by(plate, Time) %>%
summarise(
untx_mean = mean(adj_reading, na.rm = TRUE),
n_untx = n(),
.groups = "drop"
)
with_untx_75 <- processed_75 %>%
left_join(untx_per_plate_75, by = c("plate", "Time")) %>%
mutate(
norm_to_untx = if_else(
!is.na(untx_mean) & untx_mean > 0,
(adj_reading / untx_mean) * 100,
NA_real_
)
) %>%
ungroup()
wide_norm_75 <-with_untx_75 %>%
dplyr::select(Well, tx, conc, Time, plate,norm_to_untx) %>%
pivot_wider(id_cols = c(Well, tx, conc,plate), names_from = Time, values_from = norm_to_untx)
# Step 5: compute between-time ratio (two_day relative to zero) safely
final_df_75 <- wide_norm_75 %>%
mutate(
hr_norm = case_when(
is.na(T0) | is.na(T48) ~ NA_real_,
T0 == 0 ~ NA_real_, # choose NA to flag unreliable ratio (or 0 if you prefer)
TRUE ~ (T48 / T0) * 100
)
) %>%
filter(tx != "blank")final_df_75 %>%
# dplyr::rename("conc"=conc.x) %>%
ggplot(., aes(x=conc, y=hr_norm))+
geom_boxplot(aes(fill=tx))+
theme_bw()+
facet_wrap(~tx)+
ggtitle("Box plot of normalized 75 DRC")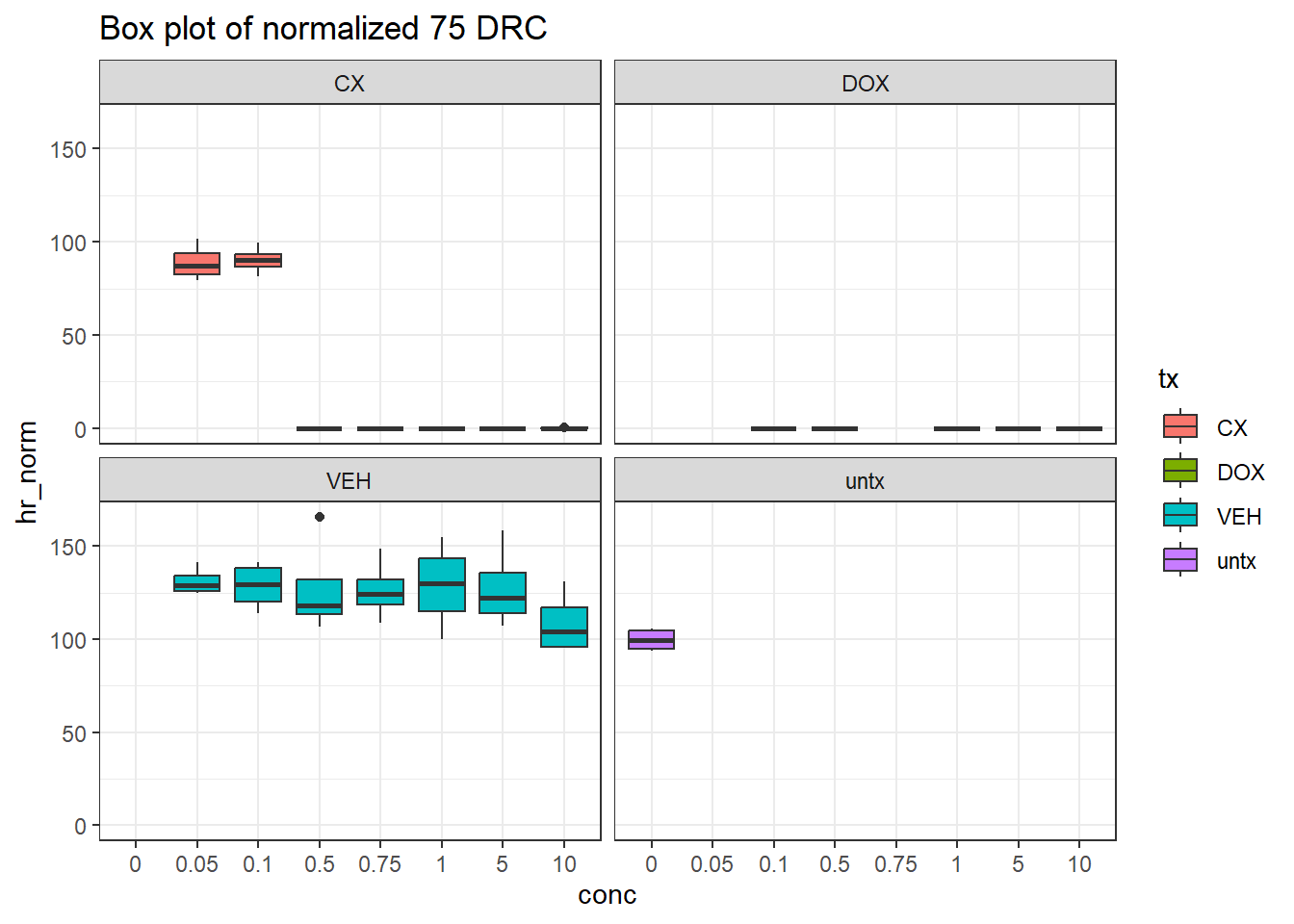
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
78-1 iPSC renee
T0_78_iPSC_p1_viability <- read_excel("data/Presto_blue_results/78_p1_iPSC_T0_251026.xls",
skip = 26)
T0_78_iPSC_p2_viability <- read_excel("data/Presto_blue_results/78_p2_iPSC_T0_251026.xls",
skip = 26)
T48_78_iPSC_p1_viability <- read_excel("data/Presto_blue_results/78_iPSC_p1_T48_251028.xlsx",
skip = 26)
T48_78_iPSC_p2_viability <- read_excel("data/Presto_blue_results/78_iPSC_p2_T48_251028.xlsx",
skip = 26)
iPSC_78_viability_data <-
T0_78_iPSC_p1_viability %>%
mutate(Well="T0_p1") %>%
bind_rows(T0_78_iPSC_p2_viability %>%
mutate(Well="T0_p2")) %>%
bind_rows(T48_78_iPSC_p1_viability %>%
mutate(Well="T48_p1")) %>%
bind_rows(T48_78_iPSC_p2_viability %>%
mutate(Well="T48_p2")) %>%
dplyr::rename("Time_Plate"=Well) %>%
pivot_longer(., A1:H12, names_to = "Well", values_to = "reading") %>%
pivot_wider(id_cols=c("Well"), names_from = "Time_Plate", values_from = "reading")iPSC_plate_map <- read_excel("data/Presto_blue_results/iPSC_cell_viability_platemap.xlsx",
skip = 9) base_df_78 <-iPSC_plate_map %>%
separate(tx_conc, into = c("tx","conc"), sep = "_") %>%
left_join(., iPSC_78_viability_data, by = "Well") %>%
pivot_longer(cols = c(T0_p1, T0_p2, T48_p1,T48_p2), names_to = "Time_plate") %>%
separate(Time_plate, into = c("Time","plate"), sep = "_") %>%
mutate(
tx = factor(tx, levels = c("CX", "DOX", "VEH", "untx", "blank")), # check these levels match your data
conc = factor(conc, levels = c("0","0.05","0.1","0.5","0.75","1","5","10","na")),
Time = factor(Time, levels = c("T0","T48"))
)
# 1. subtract blank per Time and clip negatives
processed_78 <- base_df_78 %>%
group_by(plate,Time) %>%
mutate(
blank_mean = mean(value[tx == "blank"], na.rm = TRUE),
adj_reading = pmax(value - blank_mean, 0)
) %>%
ungroup()
untx_per_plate_78 <- processed_78 %>%
filter(tx == "untx") %>%
group_by(plate, Time) %>%
summarise(
untx_mean = mean(adj_reading, na.rm = TRUE),
n_untx = n(),
.groups = "drop"
)
with_untx_78 <- processed_78 %>%
left_join(untx_per_plate_78, by = c("plate", "Time")) %>%
mutate(
norm_to_untx = if_else(
!is.na(untx_mean) & untx_mean > 0,
(adj_reading / untx_mean) * 100,
NA_real_
)
) %>%
ungroup()
wide_norm_78 <-with_untx_78 %>%
dplyr::select(Well, tx, conc, Time, plate,norm_to_untx) %>%
pivot_wider(id_cols = c(Well, tx, conc,plate), names_from = Time, values_from = norm_to_untx)
# Step 5: compute between-time ratio (two_day relative to zero) safely
final_df_78 <- wide_norm_78 %>%
mutate(
hr_norm = case_when(
is.na(T0) | is.na(T48) ~ NA_real_,
T0 == 0 ~ NA_real_, # choose NA to flag unreliable ratio (or 0 if you prefer)
TRUE ~ (T48 / T0) * 100
)
) %>%
filter(tx != "blank")final_df_78 %>%
# dplyr::rename("conc"=conc.x) %>%
ggplot(., aes(x=conc, y=hr_norm))+
geom_boxplot(aes(fill=tx))+
theme_bw()+
facet_wrap(~tx)+
ggtitle("Box plot of normalized 78 DRC")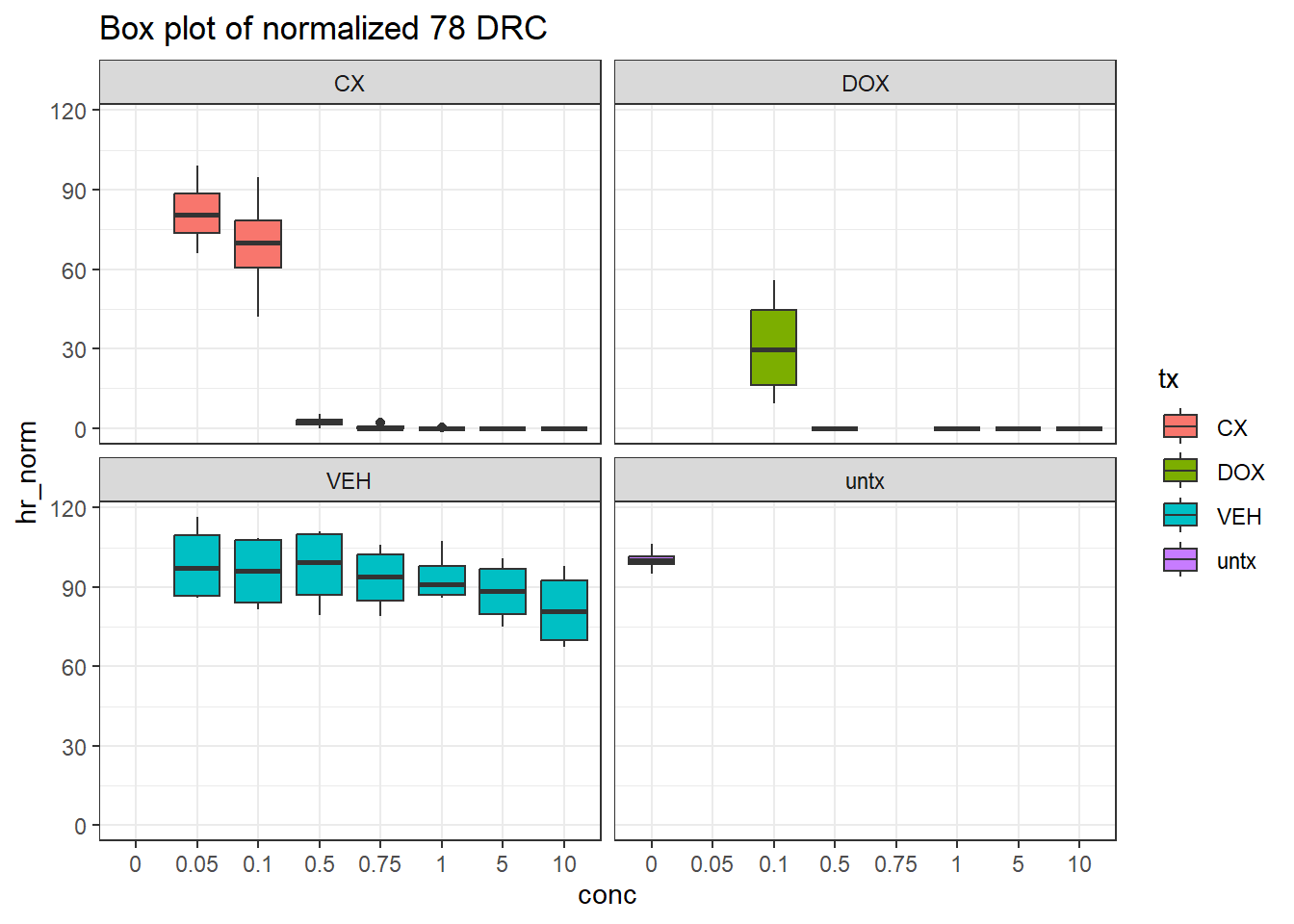
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
87-1 iPSC renee
T0_87_iPSC_p1_viability <- read_excel("data/Presto_blue_results/87_iPSC_p1_T0_251027.xlsx",
skip = 26)
T0_87_iPSC_p2_viability <- read_excel("data/Presto_blue_results/87_iPSC_p2_T0_251027.xlsx",
skip = 26)
T48_87_iPSC_p1_viability <- read_excel("data/Presto_blue_results/87_iPSC_p1_T48_251029.xlsx",
skip = 26)
T48_87_iPSC_p2_viability <- read_excel("data/Presto_blue_results/87_iPSC_p2_T48_251029.xlsx",
skip = 26)
iPSC_87_viability_data <-
T0_87_iPSC_p1_viability %>%
mutate(Well="T0_p1") %>%
bind_rows(T0_87_iPSC_p2_viability %>%
mutate(Well="T0_p2")) %>%
bind_rows(T48_87_iPSC_p1_viability %>%
mutate(Well="T48_p1")) %>%
bind_rows(T48_87_iPSC_p2_viability %>%
mutate(Well="T48_p2")) %>%
dplyr::rename("Time_Plate"=Well) %>%
pivot_longer(., A1:H12, names_to = "Well", values_to = "reading") %>%
pivot_wider(id_cols=c("Well"), names_from = "Time_Plate", values_from = "reading") base_df_87 <-iPSC_plate_map %>%
separate(tx_conc, into = c("tx","conc"), sep = "_") %>%
left_join(., iPSC_87_viability_data, by = "Well") %>%
pivot_longer(cols = c(T0_p1, T0_p2, T48_p1,T48_p2), names_to = "Time_plate") %>%
separate(Time_plate, into = c("Time","plate"), sep = "_") %>%
mutate(
tx = factor(tx, levels = c("CX", "DOX", "VEH", "untx", "blank")), # check these levels match your data
conc = factor(conc, levels = c("0","0.05","0.1","0.5","0.75","1","5","10","na")),
Time = factor(Time, levels = c("T0","T48"))
)
# 1. subtract blank per Time and clip negatives
processed_87 <- base_df_87 %>%
group_by(plate,Time) %>%
mutate(
blank_mean = mean(value[tx == "blank"], na.rm = TRUE),
adj_reading = pmax(value - blank_mean, 0)
) %>%
ungroup()
untx_per_plate_87 <- processed_87 %>%
filter(tx == "untx") %>%
group_by(plate, Time) %>%
summarise(
untx_mean = mean(adj_reading, na.rm = TRUE),
n_untx = n(),
.groups = "drop"
)
with_untx_87 <- processed_87 %>%
left_join(untx_per_plate_87, by = c("plate", "Time")) %>%
mutate(
norm_to_untx = if_else(
!is.na(untx_mean) & untx_mean > 0,
(adj_reading / untx_mean) * 100,
NA_real_
)
) %>%
ungroup()
wide_norm_87 <-with_untx_87 %>%
dplyr::select(Well, tx, conc, Time, plate,norm_to_untx) %>%
pivot_wider(id_cols = c(Well, tx, conc,plate), names_from = Time, values_from = norm_to_untx)
# Step 5: compute between-time ratio (two_day relative to zero) safely
final_df_87 <- wide_norm_87 %>%
mutate(
hr_norm = case_when(
is.na(T0) | is.na(T48) ~ NA_real_,
T0 == 0 ~ NA_real_, # choose NA to flag unreliable ratio (or 0 if you prefer)
TRUE ~ (T48 / T0) * 100
)
) %>%
filter(tx != "blank")final_df_87 %>%
# dplyr::rename("conc"=conc.x) %>%
ggplot(., aes(x=conc, y=hr_norm))+
geom_boxplot(aes(fill=tx))+
theme_bw()+
facet_wrap(~tx)+
ggtitle("Box plot of normalized 87 DRC")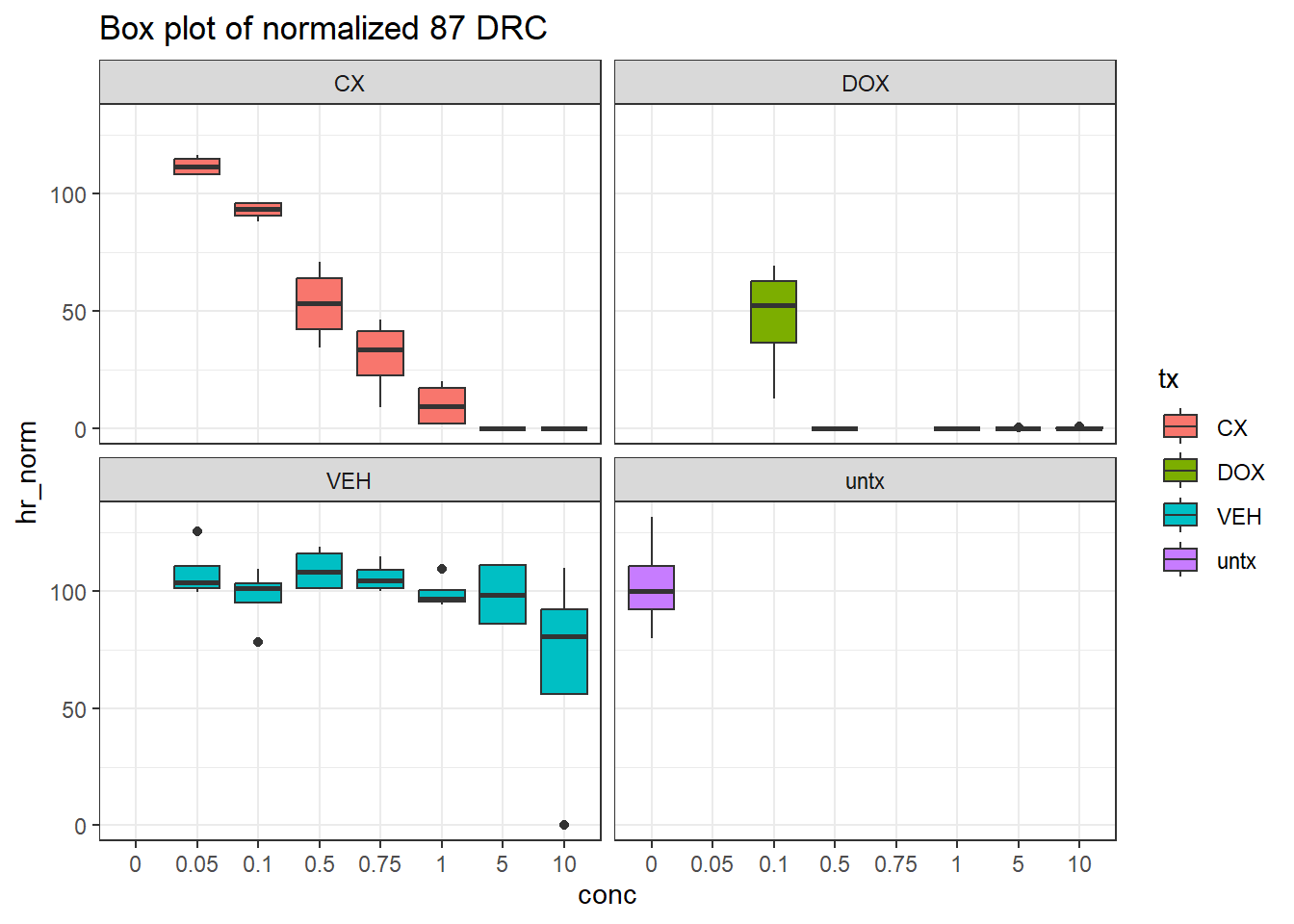
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
plot function
plot_drc <- function(data, tx_name, resp_col = "hr_norm", title = NULL, color_line = "blue") {
# Filter and prepare data
df_sub <- data %>%
filter(tx == tx_name) %>%
mutate(
conc = as.numeric(as.character(conc)),
response = as.numeric(.data[[resp_col]])
)
# Fit dose-response model
model <- drm(response ~ conc, data = df_sub, fct = L.4(fixed = c(NA, NA, 100, NA)))
# Generate log-spaced predictions
pred <- data.frame(conc = exp(seq(
log(min(df_sub$conc[df_sub$conc > 0], na.rm = TRUE)),
log(max(df_sub$conc, na.rm = TRUE)),
length.out = 100
)))
pred$response <- predict(model, newdata = pred)
# LD50
LD50 <- ED(model, 50, interval = "delta")
LD50_val <- as.numeric(LD50[1, 1])
# Create title if missing
if (is.null(title)) {
title <- paste("Dose–Response Curve for", trt_name)
}
# Plot
p <- ggplot(df_sub, aes(x = conc, y = response)) +
geom_point(size = 2, alpha = 0.8) +
geom_line(data = pred, aes(x = conc, y = response), color = color_line, size = 1) +
geom_vline(xintercept = LD50_val, color = "red", linetype = "dashed") +
annotate(
"text",
x = LD50_val,
y = max(df_sub$response, na.rm = TRUE),
label = paste0("LD50 = ", signif(LD50_val, 3)),
color = "red", angle = 90, vjust = -0.5, hjust = 1.1
) +
scale_x_log10() +
theme_minimal(base_size = 14) +
labs(title = title, x = "Concentration", y = "Response (normalized)")
# Return list so you can use both plot and model
return(list(plot = p, model = model, LD50 = LD50))
}All CX plots for iPSCs
CX_75_iPSC <- plot_drc(final_df_75, tx_name = "CX", title = "iPSC 75-1 Dose Response for CX")
Estimated effective doses
Estimate Std. Error Lower Upper
e:1:50 0.196866 0.077915 0.036398 0.357335CX_75_iPSC$plot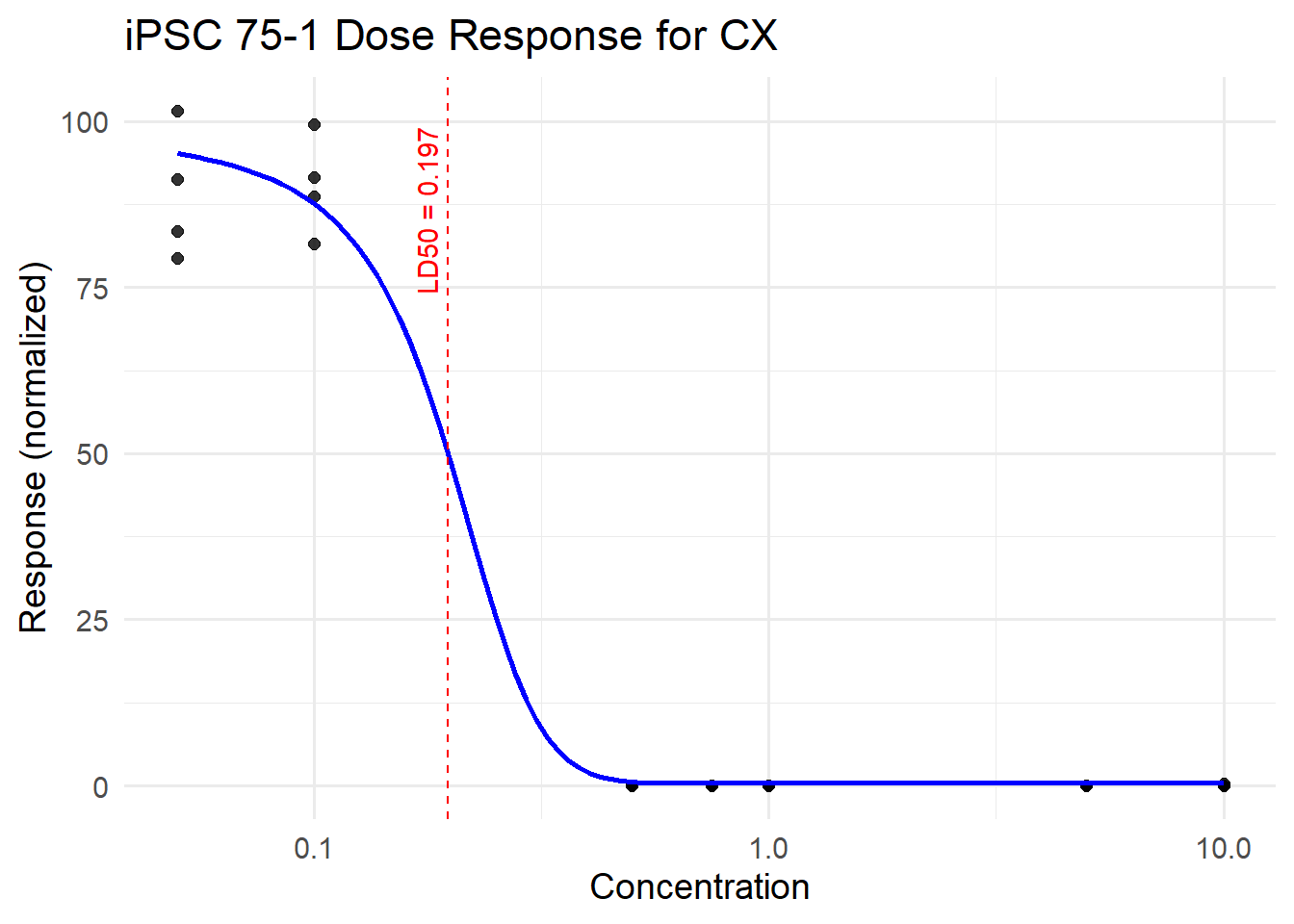
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
CX_78_iPSC <- plot_drc(final_df_78, tx_name = "CX", title = "iPSC 78-1 Dose Response for CX")
Estimated effective doses
Estimate Std. Error Lower Upper
e:1:50 0.165140 0.044385 0.073727 0.256553CX_78_iPSC$plot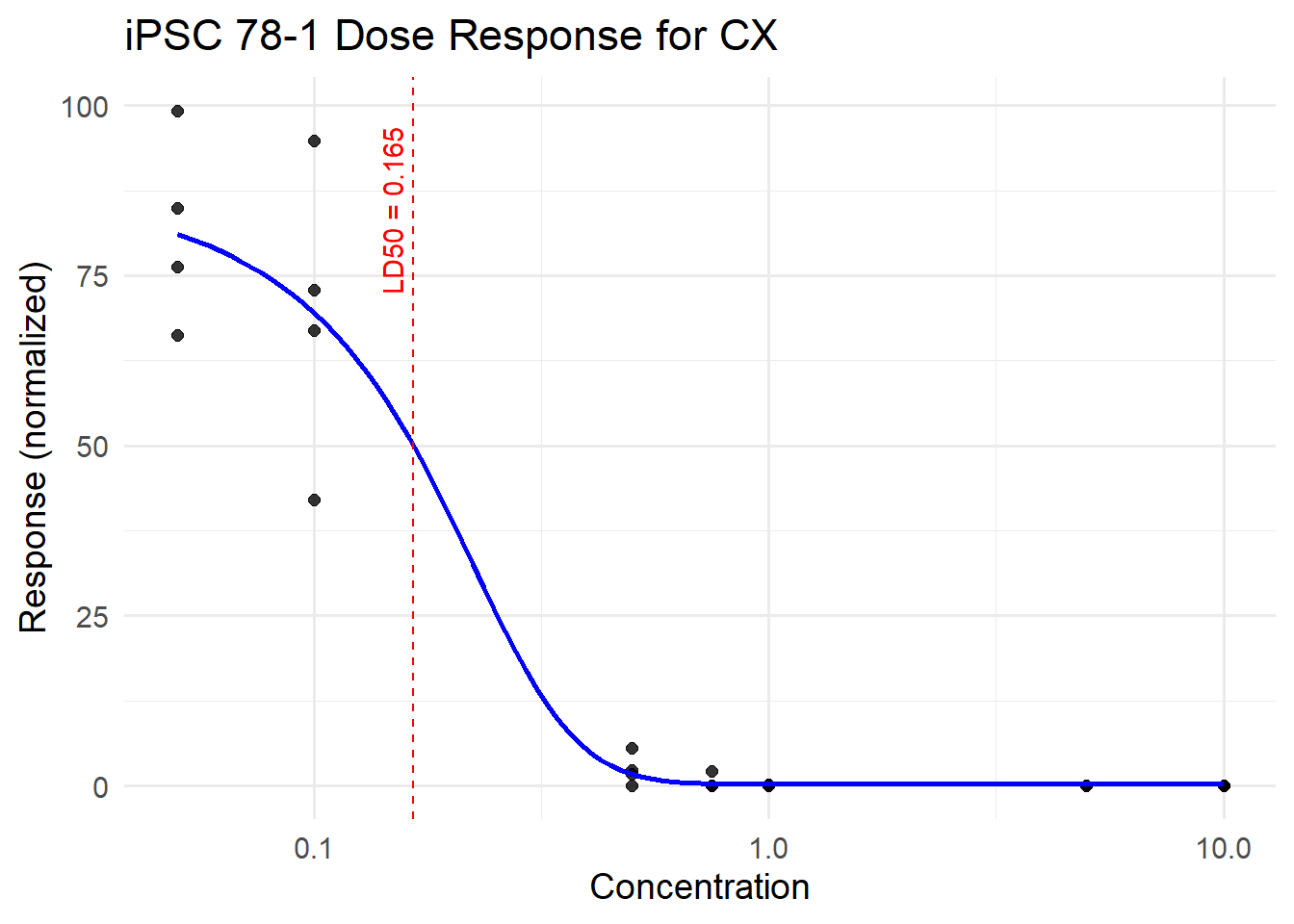
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
CX_87_iPSC <- plot_drc(final_df_87, tx_name = "CX", title = "iPSC 87-1 Dose Response for CX")
Estimated effective doses
Estimate Std. Error Lower Upper
e:1:50 0.559776 0.040025 0.477168 0.642383CX_87_iPSC$plot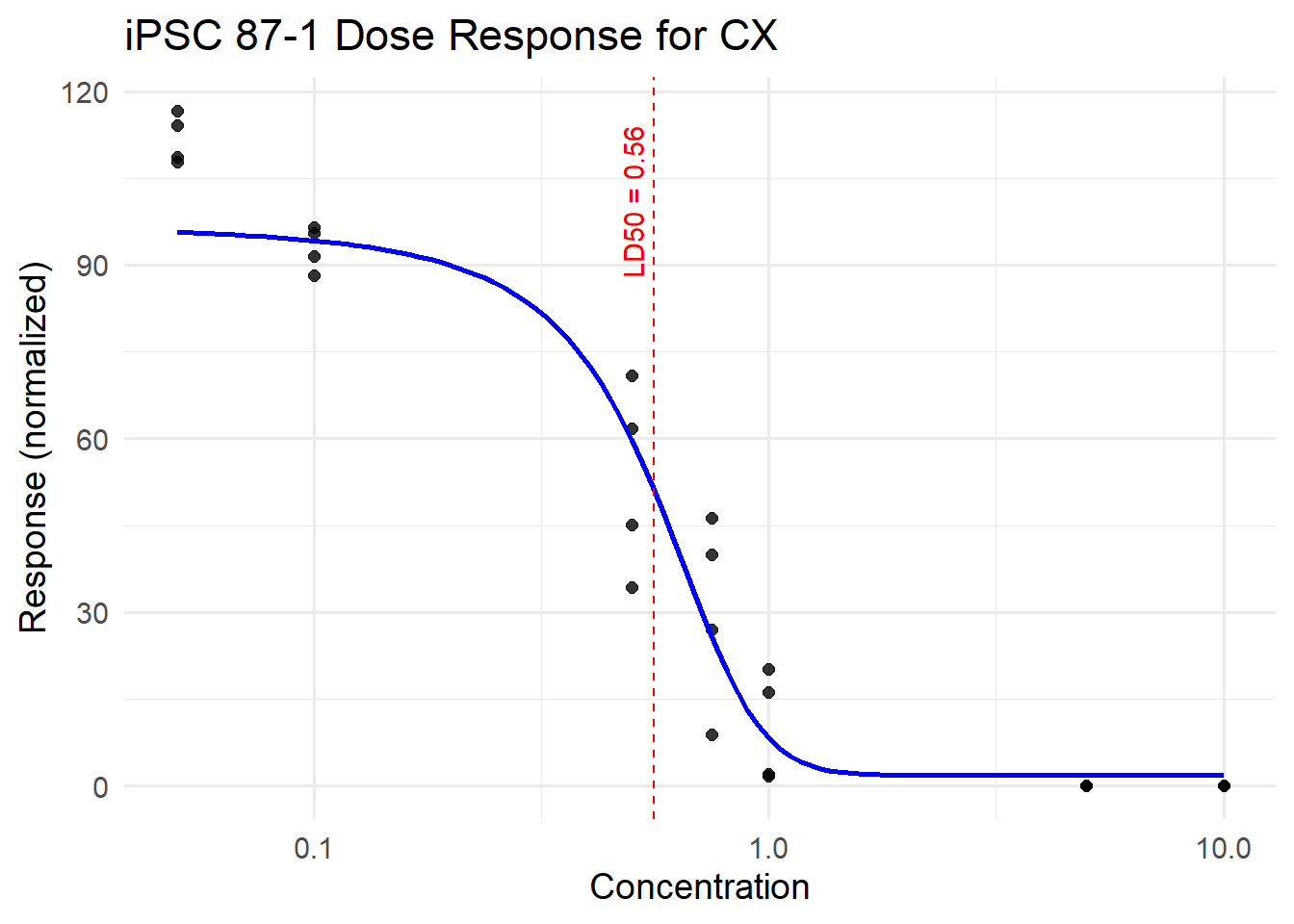
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
ggplot(
final_df_78 %>% filter(tx == "CX") %>%
mutate(conc = as.numeric(as.character(conc))),
aes(x = conc, y = hr_norm)
) +
geom_point(size = 2) +
stat_smooth(
method = drm,
method.args = list(fct = L.4(fixed = c(NA, NA, 100, NA))),
se = FALSE,
color = "blue"
) +
scale_x_log10() +
theme_minimal() +
labs(title = "CX Dose–Response", x = "Concentration", y = "Normalized viability (%)")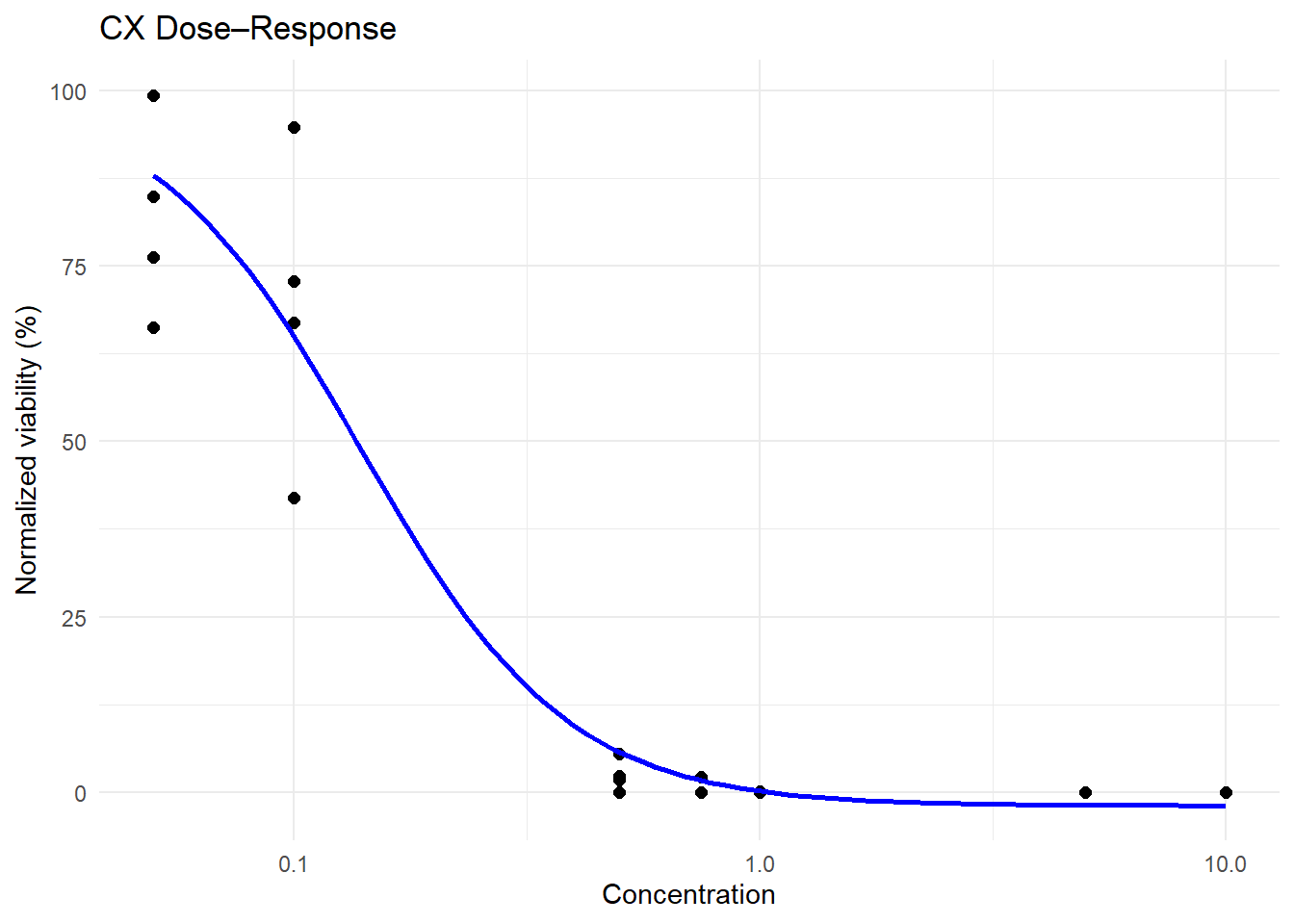
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
# write_csv(final_df_87,"data/iPSC_87_DRC_20251029.csv")
# write_csv(final_df_75,"data/iPSC_75_DRC_20251028.csv")
# write_csv(final_df_78,"data/iPSC_78_DRC_20251028.csv")Cardiomyocyte treatment
CM_platemap <- read_excel("data/Presto_blue_results/CM_cell_viability_platemap.xlsx",
skip = 9
)T0_87_CM_p1_viability <- read_excel("data/Presto_blue_results/87_iPSC-CM_p1_T0_251028.xlsx",
skip = 26)
T0_87_CM_p2_viability <- read_excel("data/Presto_blue_results/87_iPSC-CM_p2_T0_251028.xlsx",
skip = 26)
T0_87_CM_p3_viability <- read_excel("data/Presto_blue_results/87_iPSC-CM_p3_T0_251028.xlsx",
skip = 26)
T0_87_CM_p4_viability <- read_excel("data/Presto_blue_results/87_iPSC-CM_p4_T0_251028.xlsx",
skip = 26)
T3_87_CM_p1_viability <- read_excel("data/Presto_blue_results/87_iPSC-CM_p1_T3_251028.xlsx",
skip = 26)
T24_87_CM_p2_viability <- read_excel("data/Presto_blue_results/87_iPSC_CM_p2_T24_251029.xlsx",
skip = 26)
T48_87_CM_p3_viability <- read_excel("data/Presto_blue_results/87_CM_p3_T48_251030.xlsx",
skip = 26)
T96_87_CM_p4_viability <- read_excel("C:/Users/renee/Documents/Ward Lab/CX-revision/Presto Blue results/87_CM_p4_T96_251101.xlsx",
skip = 26)
CM_combo_87 <- T0_87_CM_p1_viability %>%
mutate(Well="T0_p1") %>%
bind_rows(T0_87_CM_p2_viability %>%
mutate(Well="T0_p2")) %>%
bind_rows(T0_87_CM_p3_viability %>%
mutate(Well="T0_p3")) %>%
bind_rows(T0_87_CM_p4_viability %>%
mutate(Well="T0_p4")) %>%
bind_rows(T3_87_CM_p1_viability %>%
mutate(Well="T3_p1")) %>%
bind_rows(T24_87_CM_p2_viability %>%
mutate(Well="T24_p2")) %>%
bind_rows(T48_87_CM_p3_viability %>%
mutate(Well="T48_p3")) %>%
bind_rows(T96_87_CM_p4_viability %>%
mutate(Well="T96_p4")) %>%
dplyr::rename("Time_Plate"=Well) %>%
pivot_longer(., A1:H12, names_to = "Well", values_to = "reading") %>%
pivot_wider(id_cols=c("Well"), names_from = "Time_Plate", values_from = "reading")T0_78_CM_p1_viability <- read_excel("data/Presto_blue_results/78_iPSC-CM_p1_T0_251028.xlsx",
skip = 26)
T0_78_CM_p2_viability <- read_excel("data/Presto_blue_results/78_iPSC-CM_p2_T0_251028.xlsx",
skip = 26)
T0_78_CM_p3_viability <- read_excel("data/Presto_blue_results/78_iPSC-CM_p3_T0_251028.xlsx",
skip = 26)
T0_78_CM_p4_viability <- read_excel("data/Presto_blue_results/78_iPSC-CM_p4_T0_251028.xlsx",
skip = 26)
T3_78_CM_p1_viability <- read_excel("data/Presto_blue_results/78_iPSC-CM_p1_T3_251028.xlsx",
skip = 26)
T24_78_CM_p2_viability <- read_excel("data/Presto_blue_results/78_iPSC_CM_p2_T24_251029.xlsx",
skip = 26)
T48_78_CM_p3_viability <- read_excel("data/Presto_blue_results/78_CM_p3_T48_251030.xlsx",
skip = 26)
T96_78_CM_p4_viability <- read_excel("C:/Users/renee/Documents/Ward Lab/CX-revision/Presto Blue results/78_CM_p4_T96_251101.xlsx",
skip = 26)
CM_combo_78 <- T0_78_CM_p1_viability %>%
mutate(Well="T0_p1") %>%
bind_rows(T0_78_CM_p2_viability %>%
mutate(Well="T0_p2")) %>%
bind_rows(T0_78_CM_p3_viability %>%
mutate(Well="T0_p3")) %>%
bind_rows(T0_78_CM_p4_viability %>%
mutate(Well="T0_p4")) %>%
bind_rows(T3_78_CM_p1_viability %>%
mutate(Well="T3_p1")) %>%
bind_rows(T24_78_CM_p2_viability %>%
mutate(Well="T24_p2")) %>%
bind_rows(T48_78_CM_p3_viability %>%
mutate(Well="T48_p3")) %>%
bind_rows(T96_78_CM_p4_viability %>%
mutate(Well="T96_p4")) %>%
dplyr::rename("Time_Plate"=Well) %>%
pivot_longer(., A1:H12, names_to = "Well", values_to = "reading") %>%
pivot_wider(id_cols=c("Well"), names_from = "Time_Plate", values_from = "reading")T0_75_CM_p1_viability <- read_excel("data/Presto_blue_results/75_iPSC-CM_p1_T0_251028.xlsx",
skip = 26)
T0_75_CM_p2_viability <- read_excel("data/Presto_blue_results/75_iPSC-CM_p2_T0_251028.xlsx",
skip = 26)
T0_75_CM_p3_viability <- read_excel("data/Presto_blue_results/75_iPSC-CM_p3_T0_251028.xlsx",
skip = 26)
T24_75_CM_p2_viability <- read_excel("data/Presto_blue_results/75_iPSC_CM_p2_T24_251029.xlsx",
skip = 26)
T48_75_CM_p3_viability <- read_excel("data/Presto_blue_results/75_CM_p3_T48_251030.xlsx",
skip = 26)
CM_combo_75 <- T0_75_CM_p1_viability %>%
mutate(Well="T0_p1") %>%
bind_rows(T0_75_CM_p2_viability %>%
mutate(Well="T0_p2")) %>%
bind_rows(T0_75_CM_p3_viability %>%
mutate(Well="T0_p3")) %>%
# bind_rows(T3_75_CM_p1_viability %>%
# mutate(Well="T3_p1")) %>%
bind_rows(T24_75_CM_p2_viability %>%
mutate(Well="T24_p2")) %>%
bind_rows(T48_75_CM_p3_viability %>%
mutate(Well="T48_p3")) %>%
dplyr::rename("Time_Plate"=Well) %>%
pivot_longer(., A1:H12, names_to = "Well", values_to = "reading") %>%
pivot_wider(id_cols=c("Well"), names_from = "Time_Plate", values_from = "reading")87 CM
base_df_87_CM <-
CM_platemap %>%
separate(col = tx_conc, into = c("tx", "conc"), sep = "_") %>%
left_join(., CM_combo_87, by = "Well") %>%
pivot_longer(cols = c(T0_p1:last_col()), names_to = "Time_plate") %>%
separate(Time_plate, into=c("Time","plate"), sep = "_") %>%
mutate(
tx = factor(tx, levels = c("CX", "DOX", "VEH", "untx", "blank")), # check these levels match your data
conc = factor(conc, levels = c("0","0.1","0.5","2.5","blank")),
Time = factor(Time, levels = c("T0","T3","T24","T48","T96"))
)
# 1. subtract blank per Time and clip negatives
processed_87_CM <- base_df_87_CM %>%
group_by(plate,Time) %>%
mutate(
blank_mean = mean(value[tx == "blank"], na.rm = TRUE),
adj_reading = pmax(value - blank_mean, 0)
) %>%
ungroup() %>%
##removing a well with no cells
dplyr::filter(!(plate == 3 & Well == "C7")) %>%
dplyr::filter(!(plate == 3 & Well == "C8"))
## calculate untx values to normalize all other wells
untx_per_plate_87_CM <- processed_87_CM %>%
filter(tx == "untx") %>%
group_by(plate, Time) %>%
summarise(
untx_mean = mean(adj_reading, na.rm = TRUE),
n_untx = n(),
.groups = "drop"
)
### add the untx values in the plate and normalize to untx
with_untx_87_CM <- processed_87_CM %>%
left_join(untx_per_plate_87_CM, by = c("plate", "Time")) %>%
mutate(
norm_to_untx = if_else(
!is.na(untx_mean) & untx_mean > 0,
(adj_reading / untx_mean) * 100,
NA_real_
)
) %>%
ungroup()
### Filter out blanks and pivot for T0 to T48 normalizing. (probably pivot by plate first to normalize across the times)
norm_Tx_over_T0_87_CM <- with_untx_87_CM %>%
# Keep only relevant columns
filter(tx != "blank") %>%
dplyr::select(Well, tx, conc, Time, plate, norm_to_untx) %>%
# Ensure consistent order of timepoints
mutate(Time = factor(Time, levels = unique(Time))) %>%
# Pivot wider so each timepoint is its own column
pivot_wider(
id_cols = c(Well, tx, conc, plate),
names_from = Time,
values_from = norm_to_untx
) %>%
# Pivot longer again to calculate T(x)/T0
pivot_longer(
cols = -c(Well, tx, conc, plate),
names_to = "Time",
values_to = "value"
) %>%
group_by(Well, tx, conc, plate) %>%
mutate(
T0_value = value[Time == "T0"],
Tx_over_T0 = if_else(
!is.na(T0_value) & T0_value > 0,
(value / T0_value) * 100, # convert ratio to percent
NA_real_
)
) %>%
ungroup() %>%
dplyr::select(Well, tx, conc, plate, Time, Tx_over_T0)
# norm_Tx_over_T0_87_CM <-
# with_untx_87_CM %>%
# filter(tx != "blank") %>%
# dplyr::select(Well, tx, conc, Time, plate, norm_to_untx) %>%
# pivot_wider(
# id_cols = c(Well, tx, conc, plate),
# names_from = Time,
# values_from = norm_to_untx
# ) %>%
# mutate(
# norm_T3_T0 = if_else(!is.na(T0) & T0 > 0, T3 / T0, NA_real_),
# norm_T24_T0 = if_else(!is.na(T0) & T0 > 0, T24 / T0, NA_real_))#,
# # norm_T48_T0 = if_else(!is.na(T0) & T0 > 0, T48 / T0, NA_real_),
# # norm_T96_T0 = if_else(!is.na(T0) & T0 > 0, T96 / T0, NA_real_)
# )
#
# norm_Tx_over_T0_87_CM_long <- norm_Tx_over_T0_87_CM %>%
# dplyr::select(Well, tx, conc, plate, starts_with("norm_T")) %>%
# pivot_longer(
# cols = starts_with("norm_T"),
# names_to = "norm_type",
# values_to = "Tx_over_T0"
# )plot 87
norm_Tx_over_T0_87_CM %>%
mutate(Time=factor(Time, levels = c("T0", "T3","T24","T48","T96"))) %>%
ggplot(aes(x=interaction(tx,conc),y=Tx_over_T0))+
geom_boxplot(aes(fill=Time))+
ggtitle("Treatment over time 87")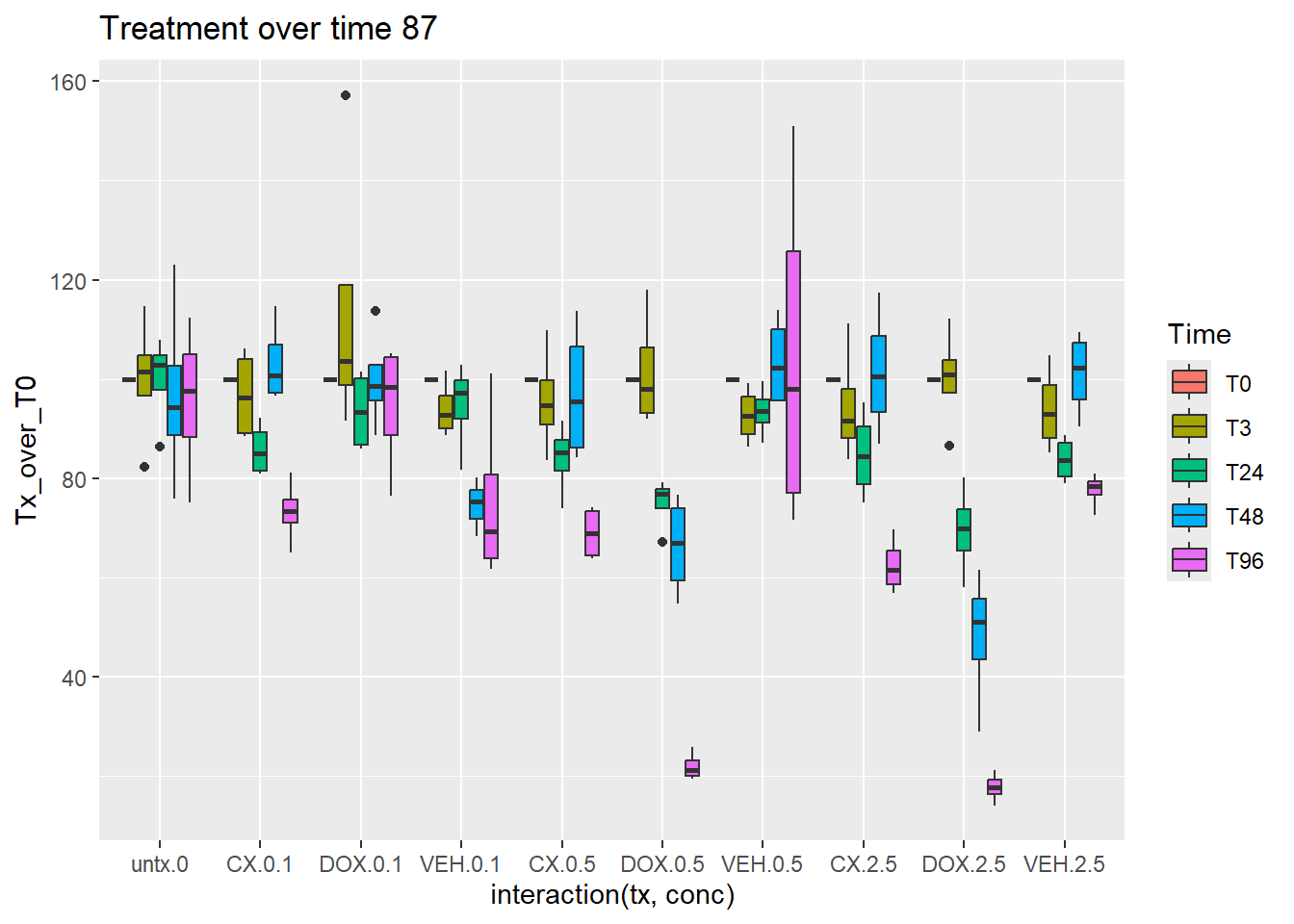
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
75 CM
base_df_75_CM <-
CM_platemap %>%
separate(col = tx_conc, into = c("tx", "conc"), sep = "_") %>%
left_join(., CM_combo_75, by = "Well") %>%
pivot_longer(cols = c(T0_p1:last_col()), names_to = "Time_plate") %>%
separate(Time_plate, into=c("Time","plate"), sep = "_") %>%
mutate(
tx = factor(tx, levels = c("CX", "DOX", "VEH", "untx", "blank")), # check these levels match your data
conc = factor(conc, levels = c("0","0.1","0.5","2.5","blank")),
Time = factor(Time, levels = c("T0","T3","T24","T48","T96"))
)
# 1. subtract blank per Time and clip negatives
processed_75_CM <- base_df_75_CM %>%
group_by(plate,Time) %>%
mutate(
blank_mean = mean(value[tx == "blank"], na.rm = TRUE),
adj_reading = pmax(value - blank_mean, 0)
) %>%
ungroup()
## calculate untx values to normalize all other wells
untx_per_plate_75_CM <- processed_75_CM %>%
filter(tx == "untx") %>%
group_by(plate, Time) %>%
summarise(
untx_mean = mean(adj_reading, na.rm = TRUE),
n_untx = n(),
.groups = "drop"
)
### add the untx values in the plate and normalize to untx
with_untx_75_CM <- processed_75_CM %>%
left_join(untx_per_plate_75_CM, by = c("plate", "Time")) %>%
mutate(
norm_to_untx = if_else(
!is.na(untx_mean) & untx_mean > 0,
(adj_reading / untx_mean) * 100,
NA_real_
)
) %>%
ungroup()
### Filter out blanks and pivot for T0 to T48 normalizing. (probably pivot by plate first to normalize across the times)
norm_Tx_over_T0_75_CM <- with_untx_75_CM %>%
# Keep only relevant columns
filter(tx != "blank") %>%
dplyr::select(Well, tx, conc, Time, plate, norm_to_untx) %>%
# Ensure consistent order of timepoints
mutate(Time = factor(Time, levels = unique(Time))) %>%
# Pivot wider so each timepoint is its own column
pivot_wider(
id_cols = c(Well, tx, conc, plate),
names_from = Time,
values_from = norm_to_untx
) %>%
# Pivot longer again to calculate T(x)/T0
pivot_longer(
cols = -c(Well, tx, conc, plate),
names_to = "Time",
values_to = "value"
) %>%
group_by(Well, tx, conc, plate) %>%
mutate(
T0_value = value[Time == "T0"],
Tx_over_T0 = if_else(
!is.na(T0_value) & T0_value > 0,
(value / T0_value) * 100, # convert ratio to percent
NA_real_
)
) %>%
ungroup() %>%
dplyr::select(Well, tx, conc, plate, Time, Tx_over_T0)
# norm_Tx_over_T0_75_CM <-
# with_untx_75_CM %>%
# filter(tx != "blank") %>%
# dplyr::select(Well, tx, conc, Time, plate, norm_to_untx) %>%
# pivot_wider(
# id_cols = c(Well, tx, conc, plate),
# names_from = Time,
# values_from = norm_to_untx
# ) %>%
# mutate(
# norm_T3_T0 = if_else(!is.na(T0) & T0 > 0, T3 / T0, NA_real_),
# norm_T24_T0 = if_else(!is.na(T0) & T0 > 0, T24 / T0, NA_real_))#,
# # norm_T48_T0 = if_else(!is.na(T0) & T0 > 0, T48 / T0, NA_real_),
# # norm_T96_T0 = if_else(!is.na(T0) & T0 > 0, T96 / T0, NA_real_)
# )
#
# norm_Tx_over_T0_75_CM_long <- norm_Tx_over_T0_75_CM %>%
# dplyr::select(Well, tx, conc, plate, starts_with("norm_T")) %>%
# pivot_longer(
# cols = starts_with("norm_T"),
# names_to = "norm_type",
# values_to = "Tx_over_T0"
# )plot 75
norm_Tx_over_T0_75_CM %>%
mutate(Time=factor(Time, levels = c("T0", "T3","T24","T48","T96"))) %>%
ggplot(aes(x=interaction(tx,conc),y=Tx_over_T0))+
geom_boxplot(aes(fill=Time))+
ggtitle("Treatment over time for 75")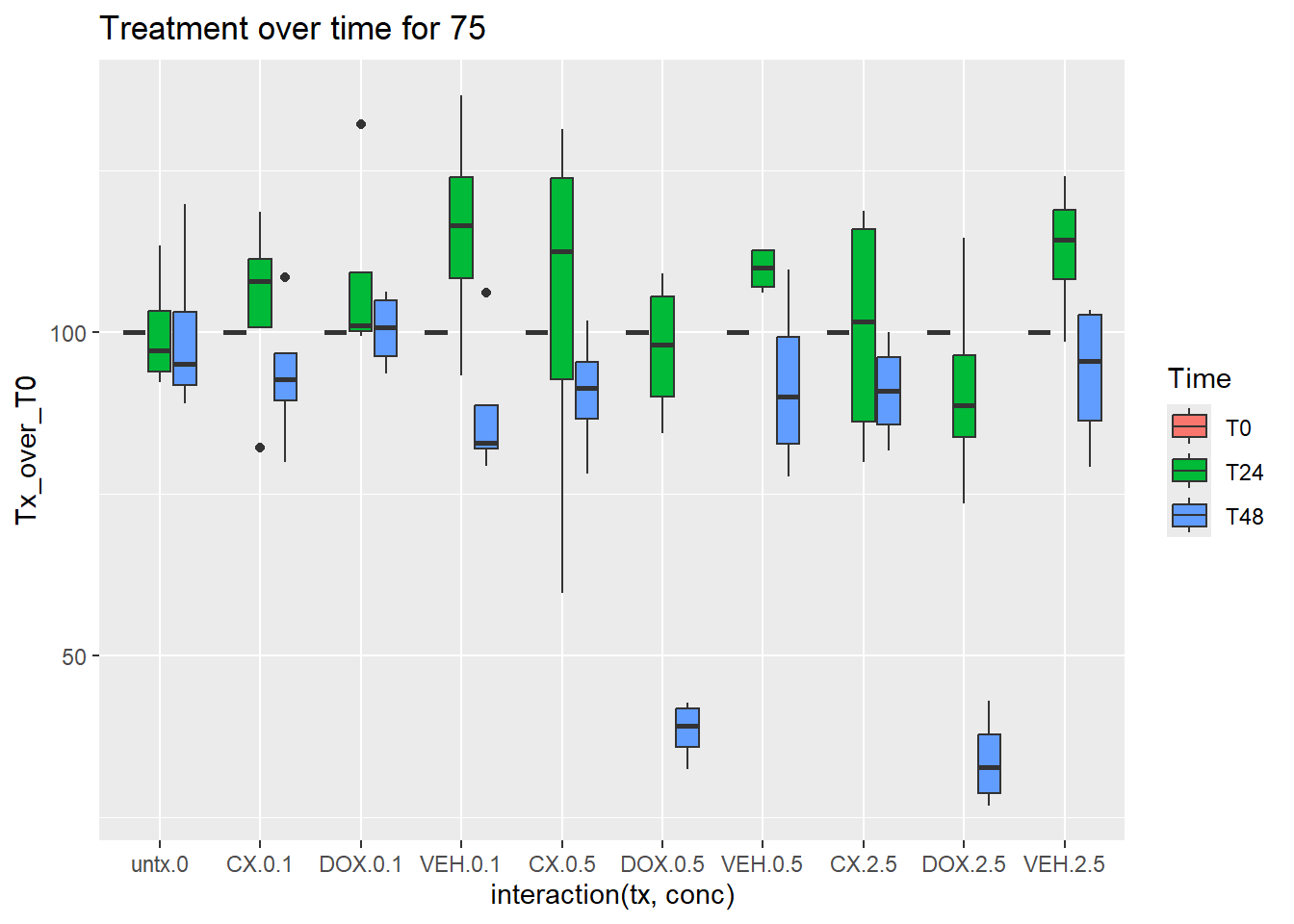
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
78 CM
base_df_78_CM <-
CM_platemap %>%
separate(col = tx_conc, into = c("tx", "conc"), sep = "_") %>%
left_join(., CM_combo_78, by = "Well") %>%
pivot_longer(cols = c(T0_p1:last_col()), names_to = "Time_plate") %>%
separate(Time_plate, into=c("Time","plate"), sep = "_") %>%
mutate(
tx = factor(tx, levels = c("CX", "DOX", "VEH", "untx", "blank")), # check these levels match your data
conc = factor(conc, levels = c("0","0.1","0.5","2.5","blank")),
Time = factor(Time, levels = c("T0","T3","T24","T48","T96"))
)
# 1. subtract blank per Time and clip negatives
processed_78_CM <- base_df_78_CM %>%
group_by(plate,Time) %>%
mutate(
blank_mean = mean(value[tx == "blank"], na.rm = TRUE),
adj_reading = pmax(value - blank_mean, 0)
) %>%
ungroup()
## calculate untx values to normalize all other wells
untx_per_plate_78_CM <- processed_78_CM %>%
filter(tx == "untx") %>%
group_by(plate, Time) %>%
summarise(
untx_mean = mean(adj_reading, na.rm = TRUE),
n_untx = n(),
.groups = "drop"
)
### add the untx values in the plate and normalize to untx
with_untx_78_CM <- processed_78_CM %>%
left_join(untx_per_plate_78_CM, by = c("plate", "Time")) %>%
mutate(
norm_to_untx = if_else(
!is.na(untx_mean) & untx_mean > 0,
(adj_reading / untx_mean) * 100,
NA_real_
)
) %>%
ungroup()
### Filter out blanks and pivot for T0 to T48 normalizing. (probably pivot by plate first to normalize across the times)
norm_Tx_over_T0_78_CM <- with_untx_78_CM %>%
# Keep only relevant columns
filter(tx != "blank") %>%
dplyr::select(Well, tx, conc, Time, plate, norm_to_untx) %>%
# Ensure consistent order of timepoints
mutate(Time = factor(Time, levels = c("T0","T3","T24","T48","T96"))) %>%
# Pivot wider so each timepoint is its own column
pivot_wider(
id_cols = c(Well, tx, conc, plate),
names_from = Time,
values_from = norm_to_untx
) %>%
# Pivot longer again to calculate T(x)/T0
pivot_longer(
cols = -c(Well, tx, conc, plate),
names_to = "Time",
values_to = "value"
) %>%
group_by(Well, tx, conc, plate) %>%
mutate(
T0_value = value[Time == "T0"],
Tx_over_T0 = if_else(
!is.na(T0_value) & T0_value > 0,
(value / T0_value) * 100, # convert ratio to percent
NA_real_
)
) %>%
ungroup() %>%
dplyr::select(Well, tx, conc, plate, Time, Tx_over_T0)plot 78
norm_Tx_over_T0_78_CM %>%
mutate(Time=factor(Time, levels = c("T0", "T3","T24","T48","T96"))) %>%
ggplot(aes(x=interaction(tx,conc),y=Tx_over_T0))+
geom_boxplot(aes(fill=Time))+
ggtitle("Treatment over time for 78")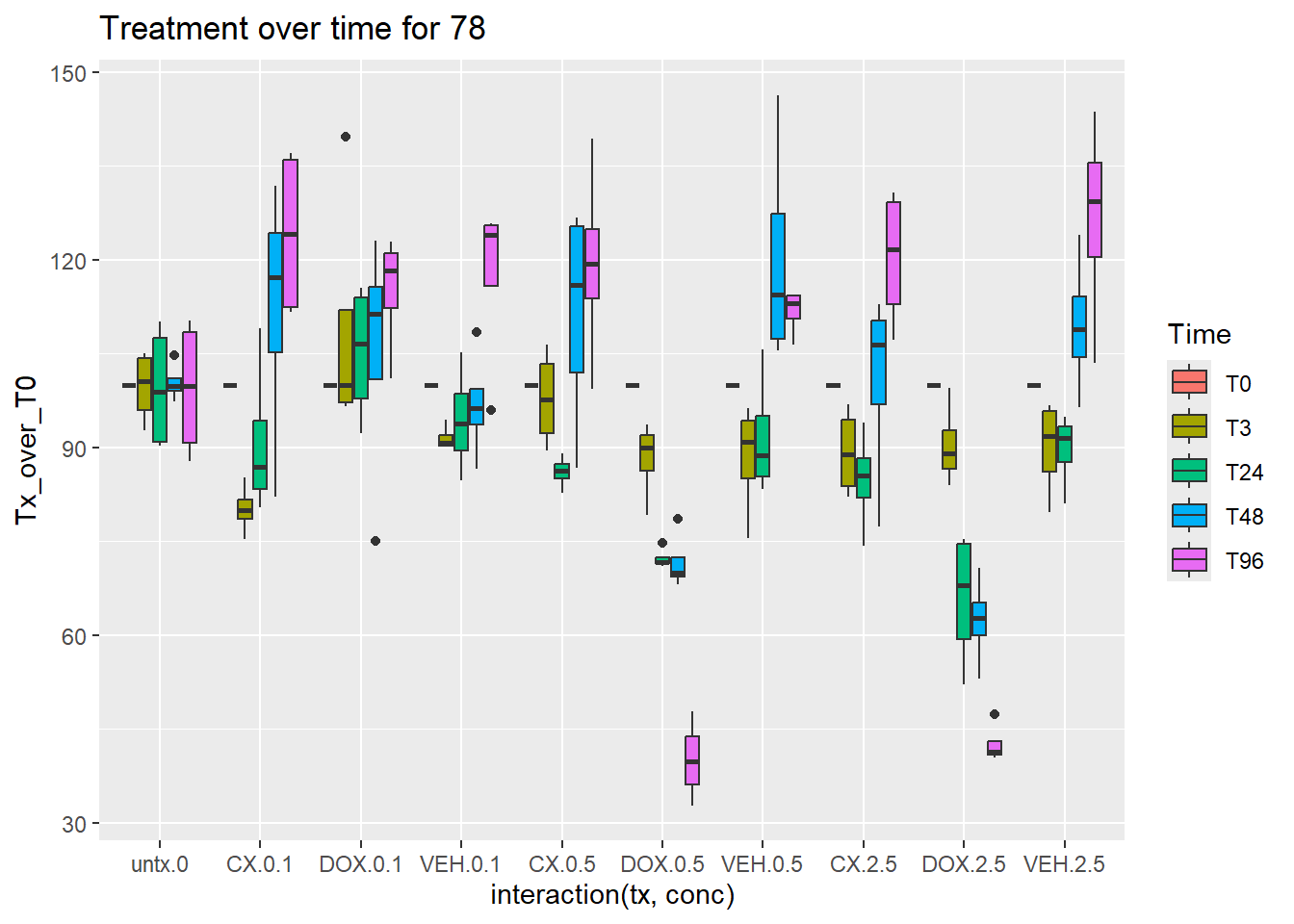
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
all_lines <- bind_rows(
norm_Tx_over_T0_87_CM %>% mutate(cell_line = "87"),
norm_Tx_over_T0_75_CM %>% mutate(cell_line = "75"),
norm_Tx_over_T0_78_CM %>% mutate(cell_line = "78")
)
summ_all <- all_lines %>%
group_by(cell_line, tx, conc, Time) %>%
summarise(
mean_Tx_over_T0 = mean(Tx_over_T0, na.rm = TRUE),
se_Tx_over_T0 = sd(Tx_over_T0, na.rm = TRUE) / sqrt(n()),
.groups = "drop"
)All normalized to untx plot
summ_all %>%
dplyr::filter(tx %in% c("CX","DOX")) %>%
mutate(Time=factor(Time, levels=c("T0","T3","T24","T48","T96")),
group=paste0(conc,"_",tx)) %>%
ggplot(aes(x=group,y=mean_Tx_over_T0))+
geom_boxplot(aes(fill=Time))+
theme_classic()+
ggtitle("Treatment over time for all lines just to untx")+
coord_cartesian(ylim = c(0,115))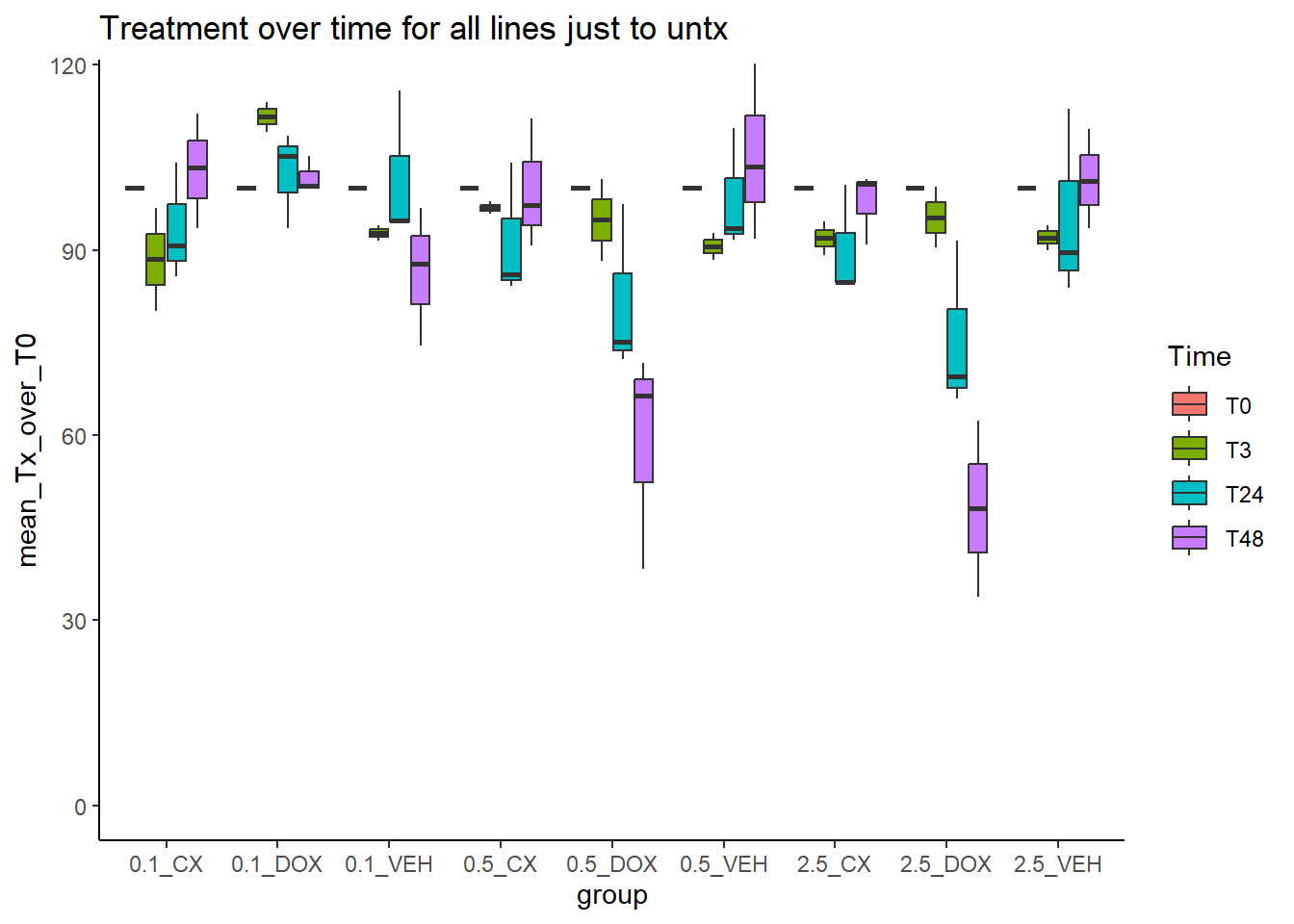
# write.csv(summ_all,"data/CM_cell_viability_3-96.csv")
# write.csv(all_lines,"data/individual_data_points_CM_cell_viability_3-96.csv")Normalizing to VEH too
normalize_to_vehicle <- function(platemap_df, combo_df) {
base_df <- platemap_df %>%
separate(col = tx_conc, into = c("tx", "conc"), sep = "_") %>%
left_join(., combo_df, by = "Well") %>%
pivot_longer(cols = c(T0_p1:last_col()), names_to = "Time_plate") %>%
separate(Time_plate, into = c("Time", "plate"), sep = "_") %>%
mutate(
tx = factor(tx, levels = c("CX", "DOX", "VEH", "untx", "blank")),
conc = factor(conc, levels = c("0","0.1","0.5","2.5","blank")),
Time = factor(Time, levels = c("T0", "T3", "T24", "T48", "T96"))
)
# 1️⃣ Subtract blank per Time and clip negatives
processed <- base_df %>%
group_by(plate, Time) %>%
mutate(
blank_mean = mean(value[tx == "blank"], na.rm = TRUE),
adj_reading = pmax(value - blank_mean, 0)
) %>%
ungroup()
# 2️⃣ Compute VEH means per plate/time/concentration
veh_per_plate <- processed %>%
dplyr::filter(tx == "VEH") %>%
group_by(plate, Time, conc) %>%
summarise(
veh_mean = mean(adj_reading, na.rm = TRUE),
n_veh = n(),
.groups = "drop"
)
# 3️⃣ Join VEH means to all wells & normalize
with_veh <- processed %>%
left_join(veh_per_plate, by = c("plate", "Time", "conc")) %>%
mutate(
norm_to_veh = if_else(
!is.na(veh_mean) & veh_mean > 0,
(adj_reading / veh_mean) * 100,
NA_real_
)
) %>%
ungroup()
# 4️⃣ Normalize each timepoint to T0 (Tx/T0)
norm_Tx_over_T0 <- with_veh %>%
dplyr::filter(tx != "blank") %>%
dplyr::select(Well, tx, conc, Time, plate, norm_to_veh) %>%
mutate(Time = factor(Time, levels = c("T0", "T3", "T24", "T48"))) %>%
pivot_wider(
id_cols = c(Well, tx, conc, plate),
names_from = Time,
values_from = norm_to_veh
) %>%
pivot_longer(
cols = -c(Well, tx, conc, plate),
names_to = "Time",
values_to = "value"
) %>%
group_by(Well, tx, conc, plate) %>%
mutate(
T0_value = value[Time == "T0"],
Tx_over_T0 = if_else(
!is.na(T0_value) & T0_value > 0,
(value / T0_value) * 100,
NA_real_
)
) %>%
ungroup() %>%
dplyr::select(Well, tx, conc, plate, Time, Tx_over_T0)
return(norm_Tx_over_T0)
}VEH_norm_plot_CM_75 <- normalize_to_vehicle(CM_platemap,CM_combo_75)
VEH_norm_plot_CM_78 <- normalize_to_vehicle(CM_platemap,CM_combo_78)
VEH_norm_plot_CM_87 <- normalize_to_vehicle(CM_platemap,CM_combo_87)VEH normed plot
all_lines_VEH_norm <- bind_rows(
VEH_norm_plot_CM_87 %>% mutate(cell_line = "87"),
VEH_norm_plot_CM_75 %>% mutate(cell_line = "75"),
VEH_norm_plot_CM_78 %>% mutate(cell_line = "78")
)
summ_all_VEH_norm <- all_lines_VEH_norm %>%
group_by(cell_line, tx, conc, Time) %>%
summarise(
mean_Tx_over_T0 = mean(Tx_over_T0, na.rm = TRUE),
se_Tx_over_T0 = sd(Tx_over_T0, na.rm = TRUE) / sqrt(n()),
.groups = "drop"
)summ_all_VEH_norm %>%
dplyr::filter(tx %in% c("CX","DOX","VEH")) %>%
mutate(Time=factor(Time, levels=c("T0","T3","T24","T48")),
group=paste0(conc,"_",tx)) %>%
ggplot(aes(x=group,y=mean_Tx_over_T0))+
geom_boxplot(aes(fill=Time))+
theme_classic()+
ggtitle("Treatment over time for all lines VEH norm")+
coord_cartesian(ylim = c(0,125))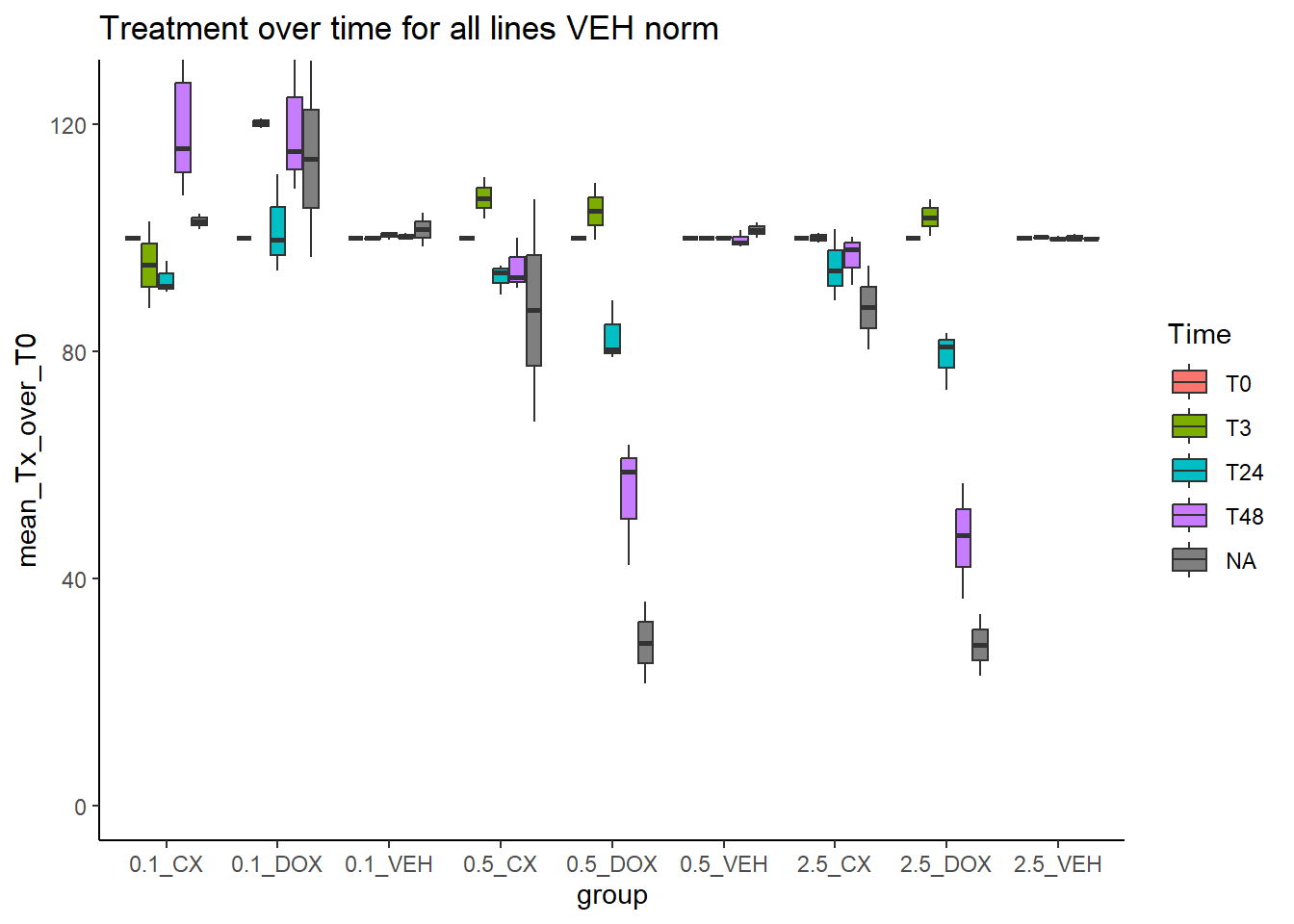
| Version | Author | Date |
|---|---|---|
| f2ba629 | reneeisnowhere | 2025-10-31 |
sessionInfo()R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 26100)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] drc_3.0-1 MASS_7.3-65 readxl_1.4.5 lubridate_1.9.4
[5] forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4 purrr_1.1.0
[9] readr_2.1.5 tidyr_1.3.1 tibble_3.3.0 ggplot2_3.5.2
[13] tidyverse_2.0.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] gtable_0.3.6 xfun_0.52 bslib_0.9.0 processx_3.8.6
[5] lattice_0.22-7 callr_3.7.6 tzdb_0.5.0 vctrs_0.6.5
[9] tools_4.4.2 ps_1.9.1 generics_0.1.4 sandwich_3.1-1
[13] pkgconfig_2.0.3 Matrix_1.7-3 RColorBrewer_1.1-3 lifecycle_1.0.4
[17] compiler_4.4.2 farver_2.1.2 git2r_0.36.2 codetools_0.2-20
[21] getPass_0.2-4 carData_3.0-5 httpuv_1.6.16 htmltools_0.5.8.1
[25] sass_0.4.10 yaml_2.3.10 Formula_1.2-5 later_1.4.2
[29] pillar_1.11.0 car_3.1-3 jquerylib_0.1.4 whisker_0.4.1
[33] cachem_1.1.0 abind_1.4-8 multcomp_1.4-28 nlme_3.1-168
[37] gtools_3.9.5 tidyselect_1.2.1 digest_0.6.37 mvtnorm_1.3-3
[41] stringi_1.8.7 labeling_0.4.3 splines_4.4.2 rprojroot_2.1.1
[45] fastmap_1.2.0 grid_4.4.2 cli_3.6.5 magrittr_2.0.3
[49] survival_3.8-3 dichromat_2.0-0.1 TH.data_1.1-4 withr_3.0.2
[53] scales_1.4.0 promises_1.3.3 plotrix_3.8-4 timechange_0.3.0
[57] rmarkdown_2.29 httr_1.4.7 cellranger_1.1.0 zoo_1.8-14
[61] hms_1.1.3 evaluate_1.0.5 knitr_1.50 mgcv_1.9-3
[65] rlang_1.1.6 Rcpp_1.1.0 glue_1.8.0 rstudioapi_0.17.1
[69] jsonlite_2.0.0 R6_2.6.1 fs_1.6.6